Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity
- PMID: 29752062
- PMCID: PMC5971117
- DOI: 10.1016/j.immuni.2018.04.006
Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity
Abstract
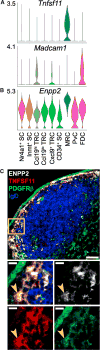
Stromal cells (SCs) establish the compartmentalization of lymphoid tissues critical to the immune response. However, the full diversity of lymph node (LN) SCs remains undefined. Using droplet-based single-cell RNA sequencing, we identified nine peripheral LN non-endothelial SC clusters. Included are the established subsets, Ccl19hi T-zone reticular cells (TRCs), marginal reticular cells, follicular dendritic cells (FDCs), and perivascular cells. We also identified Ccl19lo TRCs, likely including cholesterol-25-hydroxylase+ cells located at the T-zone perimeter, Cxcl9+ TRCs in the T-zone and interfollicular region, CD34+ SCs in the capsule and medullary vessel adventitia, indolethylamine N-methyltransferase+ SCs in the medullary cords, and Nr4a1+ SCs in several niches. These data help define how transcriptionally distinct LN SCs support niche-restricted immune functions and provide evidence that many SCs are in an activated state.
Keywords: double negative cell; fibroblastic reticular cell; follicular dendritic cell; marginal reticular cell; perivascular cell; single-cell RNA sequencing; stromal cell.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Allen CDC, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. - PubMed
-
- Ara T, Tokoyoda K, Sugiyama T, Egawa T, Kawabata K, Nagasawa T. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity. 2003;19:257–267. - PubMed
-
- Armulik A, Genové G, Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell. 2011;21:193–215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

