Functional analysis of Hsh155/SF3b1 interactions with the U2 snRNA/branch site duplex
- PMID: 29752352
- PMCID: PMC6049509
- DOI: 10.1261/rna.065664.118
Functional analysis of Hsh155/SF3b1 interactions with the U2 snRNA/branch site duplex
Abstract
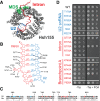
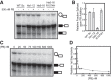
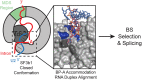
SF3b1 is an essential component of the U2 snRNP implicated in branch site (BS) recognition and found to be frequently mutated in several human cancers. While recent structures of yeast and human SF3b1 have revealed its molecular architecture, the importance of specific RNA:protein contacts and conformational changes remains largely uncharacterized. Here, we performed mutational analysis of yeast SF3b1, guided by recent structures of the spliceosome. We find that conserved amino acids contacting the U2 snRNA backbone of the U2/BS duplex are nonessential, and that yeast can tolerate truncation of the HEAT repeats containing these amino acids. The pocket housing the branchpoint adenosine (BP-A) is also amenable to mutation despite strong conservation. However, mutations that support viability can still lead to defects in splicing pre-mRNAs with nonconsensus BS substitutions found at -3, -2, -1, and +1 positions relative to the BP-A or at the branchpoint position. Through the generation of yeast and human chimeric proteins, we further defined the functionally conserved regions of Hsh155 as well as identify changes in BS usage resulting from inclusion of human SF3b1 HEAT repeats. Moreover, these chimeric proteins confer a sensitivity to small molecule inhibition by pladienolide B to yeast splicing. Together, these data reveal the importance of individual contacts of Hsh155/SF3b1 to the U2/BS duplex and define their contribution to BS usage by the spliceosome.
Keywords: SF3b1; pladienolide B; snRNP; spliceosome; splicing.
© 2018 Carrocci et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
-
- Agrawal AA, Yu L, Smith PG, Buonamici S. 2017. Targeting splicing abnormalities in cancer. Curr Opin Genet Dev 48: 67–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
