Mutations in PPCS, Encoding Phosphopantothenoylcysteine Synthetase, Cause Autosomal-Recessive Dilated Cardiomyopathy
- PMID: 29754768
- PMCID: PMC5992122
- DOI: 10.1016/j.ajhg.2018.03.022
Mutations in PPCS, Encoding Phosphopantothenoylcysteine Synthetase, Cause Autosomal-Recessive Dilated Cardiomyopathy
Abstract
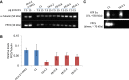
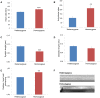
Coenzyme A (CoA) is an essential metabolic cofactor used by around 4% of cellular enzymes. Its role is to carry and transfer acetyl and acyl groups to other molecules. Cells can synthesize CoA de novo from vitamin B5 (pantothenate) through five consecutive enzymatic steps. Phosphopantothenoylcysteine synthetase (PPCS) catalyzes the second step of the pathway during which phosphopantothenate reacts with ATP and cysteine to form phosphopantothenoylcysteine. Inborn errors of CoA biosynthesis have been implicated in neurodegeneration with brain iron accumulation (NBIA), a group of rare neurological disorders characterized by accumulation of iron in the basal ganglia and progressive neurodegeneration. Exome sequencing in five individuals from two unrelated families presenting with dilated cardiomyopathy revealed biallelic mutations in PPCS, linking CoA synthesis with a cardiac phenotype. Studies in yeast and fruit flies confirmed the pathogenicity of identified mutations. Biochemical analysis revealed a decrease in CoA levels in fibroblasts of all affected individuals. CoA biosynthesis can occur with pantethine as a source independent from PPCS, suggesting pantethine as targeted treatment for the affected individuals still alive.
Keywords: PPCS; coenzyme A; dilated cardiomyopathy; pantethine treatment; pentothenate; phospohopantothenoylcysteine synthetase.
Copyright © 2018 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Hartig M.B., Hörtnagel K., Garavaglia B., Zorzi G., Kmiec T., Klopstock T., Rostasy K., Svetel M., Kostic V.S., Schuelke M. Genotypic and phenotypic spectrum of PANK2 mutations in patients with neurodegeneration with brain iron accumulation. Ann. Neurol. 2006;59:248–256. - PubMed
-
- Annesi G., Gagliardi M., Iannello G., Quattrone A., Iannello G., Quattrone A. Mutational analysis of COASY in an Italian patient with NBIA. Parkinsonism Relat. Disord. 2016;28:150–151. - PubMed
-
- Evers C., Seitz A., Assmann B., Opladen T., Karch S., Hinderhofer K., Granzow M., Paramasivam N., Eils R., Diessl N. Diagnosis of CoPAN by whole exome sequencing: Waking up a sleeping tiger’s eye. Am. J. Med. Genet. A. 2017;173:1878–1886. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

