Polε Instability Drives Replication Stress, Abnormal Development, and Tumorigenesis
- PMID: 29754823
- PMCID: PMC5972231
- DOI: 10.1016/j.molcel.2018.04.008
Polε Instability Drives Replication Stress, Abnormal Development, and Tumorigenesis
Abstract
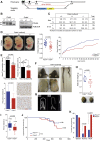
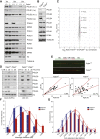
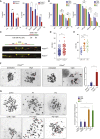
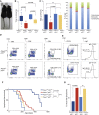
DNA polymerase ε (POLE) is a four-subunit complex and the major leading strand polymerase in eukaryotes. Budding yeast orthologs of POLE3 and POLE4 promote Polε processivity in vitro but are dispensable for viability in vivo. Here, we report that POLE4 deficiency in mice destabilizes the entire Polε complex, leading to embryonic lethality in inbred strains and extensive developmental abnormalities, leukopenia, and tumor predisposition in outbred strains. Comparable phenotypes of growth retardation and immunodeficiency are also observed in human patients harboring destabilizing mutations in POLE1. In both Pole4-/- mouse and POLE1 mutant human cells, Polε hypomorphy is associated with replication stress and p53 activation, which we attribute to inefficient replication origin firing. Strikingly, removing p53 is sufficient to rescue embryonic lethality and all developmental abnormalities in Pole4 null mice. However, Pole4-/-p53+/- mice exhibit accelerated tumorigenesis, revealing an important role for controlled CMG and origin activation in normal development and tumor prevention.
Keywords: DNA polymerase ε; DNA replication; POLE1/2 mutations; genome stability; p53; tumorigenesis.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. - PMC - PubMed
-
- Bellelli R., Castellone M.D., Guida T., Limongello R., Dathan N.A., Merolla F., Cirafici A.M., Affuso A., Masai H., Costanzo V. NCOA4 transcriptional coactivator inhibits activation of DNA replication origins. Mol. Cell. 2014;55:123–137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

