Medicago truncatula SOC1 Genes Are Up-regulated by Environmental Cues That Promote Flowering
- PMID: 29755488
- PMCID: PMC5934494
- DOI: 10.3389/fpls.2018.00496
Medicago truncatula SOC1 Genes Are Up-regulated by Environmental Cues That Promote Flowering
Abstract
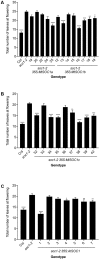
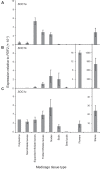
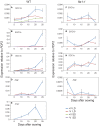
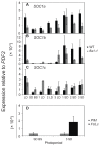
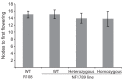
Like Arabidopsis thaliana, the flowering of the legume Medicago truncatula is promoted by long day (LD) photoperiod and vernalization. However, there are differences in the molecular mechanisms involved, with orthologs of two key Arabidopsis thaliana regulators, FLOWERING LOCUS C (FLC) and CONSTANS (CO), being absent or not having a role in flowering time function in Medicago. In Arabidopsis, the MADS-box transcription factor gene, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (AtSOC1), plays a key role in integrating the photoperiodic and vernalization pathways. In this study, we set out to investigate whether the Medicago SOC1 genes play a role in regulating flowering time. Three Medicago SOC1 genes were identified and characterized (MtSOC1a-MtSOC1c). All three MtSOC1 genes, when heterologously expressed, were able to promote earlier flowering of the late-flowering Arabidopsis soc1-2 mutant. The three MtSOC1 genes have different patterns of expression. However, consistent with a potential role in flowering time regulation, all three MtSOC1 genes are expressed in the shoot apex and are up-regulated in the shoot apex of plants in response to LD photoperiods and vernalization. The up-regulation of MtSOC1 genes was reduced in Medicago fta1-1 mutants, indicating that they are downstream of MtFTa1. Insertion mutant alleles of Medicago soc1b do not flower late, suggestive of functional redundancy among Medicago SOC1 genes in promoting flowering.
Keywords: Medicago; flowering time; genome evolution; legume; photoperiod; vernalization.
Figures
Similar articles
-
Mechanisms of Vernalization-Induced Flowering in Legumes.Int J Mol Sci. 2022 Aug 31;23(17):9889. doi: 10.3390/ijms23179889. Int J Mol Sci. 2022. PMID: 36077286 Free PMC article. Review.
-
Medicago PHYA promotes flowering, primary stem elongation and expression of flowering time genes in long days.BMC Plant Biol. 2020 Jul 11;20(1):329. doi: 10.1186/s12870-020-02540-y. BMC Plant Biol. 2020. PMID: 32652925 Free PMC article.
-
Gene-edited Mtsoc1 triple mutant Medicago plants do not flower.Front Plant Sci. 2024 Feb 26;15:1357924. doi: 10.3389/fpls.2024.1357924. eCollection 2024. Front Plant Sci. 2024. PMID: 38469328 Free PMC article.
-
The Candidate Photoperiod Gene MtFE Promotes Growth and Flowering in Medicago truncatula.Front Plant Sci. 2021 Mar 26;12:634091. doi: 10.3389/fpls.2021.634091. eCollection 2021. Front Plant Sci. 2021. PMID: 33841463 Free PMC article.
-
Regulation and function of SOC1, a flowering pathway integrator.J Exp Bot. 2010 May;61(9):2247-54. doi: 10.1093/jxb/erq098. Epub 2010 Apr 22. J Exp Bot. 2010. PMID: 20413527 Review.
Cited by
-
MtING2 encodes an ING domain PHD finger protein which affects Medicago growth, flowering, global patterns of H3K4me3, and gene expression.Plant J. 2022 Nov;112(4):1029-1050. doi: 10.1111/tpj.15994. Epub 2022 Oct 17. Plant J. 2022. PMID: 36178149 Free PMC article.
-
Mechanisms of Vernalization-Induced Flowering in Legumes.Int J Mol Sci. 2022 Aug 31;23(17):9889. doi: 10.3390/ijms23179889. Int J Mol Sci. 2022. PMID: 36077286 Free PMC article. Review.
-
The SOC1-like gene BoMADS50 is associated with the flowering of Bambusa oldhamii.Hortic Res. 2021 Jun 1;8(1):133. doi: 10.1038/s41438-021-00557-4. Hortic Res. 2021. PMID: 34059654 Free PMC article.
-
Medicago PHYA promotes flowering, primary stem elongation and expression of flowering time genes in long days.BMC Plant Biol. 2020 Jul 11;20(1):329. doi: 10.1186/s12870-020-02540-y. BMC Plant Biol. 2020. PMID: 32652925 Free PMC article.
-
The transcriptomic response to a short day to long day shift in leaves of the reference legume Medicago truncatula.PeerJ. 2019 Mar 22;7:e6626. doi: 10.7717/peerj.6626. eCollection 2019. PeerJ. 2019. PMID: 30923654 Free PMC article.
References
-
- Benlloch R., d’Erfurth I., Ferrandiz C., Cosson V., Beltran J. P., Canas L. A., et al. (2006). Isolation of MtPIM proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol. 142 972–983. 10.1104/pp.106.083543 - DOI - PMC - PubMed
-
- Bertioli D. J., Moretzsohn M. C., Madsen L. H., Sandal N., Leal-Bertioli S. C., Guimaraes P. M., et al. (2009). An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, genomes stability and evolution of legume. BMC Genomics 10:45. 10.1186/1471-2164-10-45 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

