Functional Conservation and Divergence of Soybean GmSTOP1 Members in Proton and Aluminum Tolerance
- PMID: 29755502
- PMCID: PMC5932199
- DOI: 10.3389/fpls.2018.00570
Functional Conservation and Divergence of Soybean GmSTOP1 Members in Proton and Aluminum Tolerance
Abstract
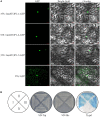
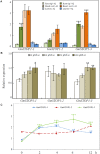
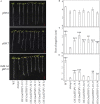
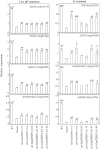
Proton (H+) and aluminum (Al) rhizotoxicity are two major factors limiting crop production in acid soils. Orthologs of the zinc-finger transcription factor, Sensitive To Proton Rhizotoxicity1 (STOP1), have been found to play an essential role in the tolerance to both stresses by regulating the transcription of multiple H+ and Al tolerant genes. In the present study, color three GmSTOP1 homologs were identified in the soybean genome. All three GmSTOP1 exhibited similar properties as reflected by the harboring of four potential zinc finger domains, localizing in the nucleus, and having transactivation activity. Expression profiling showed that H+ stress slightly modulated transcription of all three GmSTOP1s, while Al significantly up-regulated GmSTOP1-1 and GmSTOP1-3 in root apexes and GmSTOP1-3 in basal root regions. Furthermore, complementation assays in an Arabidopsis Atstop1 mutant line overexpressing these GmSTOP1s demonstrated that all three GmSTOP1s largely reverse the H+ sensitivity of the Atstop1 mutant and restore the expression of genes involved in H+ tolerance. In contrast, only GmSTOP1-1 and GmSTOP1-3 could partially recover Al tolerance in the Atstop1 mutant. These results suggest that the function of three GmSTOP1s is evolutionarily conserved in H+ tolerance, but not in Al tolerance.
Keywords: Al rhizotoxicity; GmSTOP1; proton rhizotoxicity; soybean; transcription factor.
Figures

References
-
- Badia M. B., Arias C. L., Tronconi M. A., Maurino V. G., Andreo C. S., Drincovich M. F., et al. (2015). Enhanced cytosolic NADP-ME2 activity in A. thaliana affects plant development, stress tolerance and specific diurnal and nocturnal cellular processes. Plant Sci. 240 193–203. 10.1016/j.plantsci.2015.09.015 - DOI - PubMed
-
- Chang Y. C., Yamamoto Y., Matsumoto H. (1999). Accumulation of aluminium in the cell wall pectin in cultured tobacco (Nicotiana tabacum L.) cells treated with a combination of aluminium and iron. Plant Cell Environ. 22 1009–1017. 10.1046/j.1365-3040.1999.00467.x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

