Bacterial CRISPR Regions: General Features and their Potential for Epidemiological Molecular Typing Studies
- PMID: 29755603
- PMCID: PMC5925864
- DOI: 10.2174/1874285801812010059
Bacterial CRISPR Regions: General Features and their Potential for Epidemiological Molecular Typing Studies
Abstract
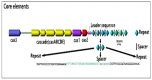
Introduction: CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) loci as novel and applicable regions in prokaryotic genomes have gained great attraction in the post genomics era.
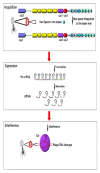
Methods: These unique regions are diverse in number and sequence composition in different pathogenic bacteria and thereby can be a suitable candidate for molecular epidemiology and genotyping studies. Results:Furthermore, the arrayed structure of CRISPR loci (several unique repeats spaced with the variable sequence) and associated cas genes act as an active prokaryotic immune system against viral replication and conjugative elements. This property can be used as a tool for RNA editing in bioengineering studies.
Conclusion: The aim of this review was to survey some details about the history, nature, and potential applications of CRISPR arrays in both genetic engineering and bacterial genotyping studies.
Keywords: Bioengineering; CRISPR; Epidemiological studies; Genotyping; Pathogenic bacteria; Prokaryotic immune system.
Similar articles
-
Survey of clustered regularly interspaced short palindromic repeats and their associated Cas proteins (CRISPR/Cas) systems in multiple sequenced strains of Klebsiella pneumoniae.BMC Res Notes. 2015 Aug 4;8:332. doi: 10.1186/s13104-015-1285-7. BMC Res Notes. 2015. PMID: 26238567 Free PMC article.
-
The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes.mBio. 2017 Sep 19;8(5):e01397-17. doi: 10.1128/mBio.01397-17. mBio. 2017. PMID: 28928211 Free PMC article.
-
Lactobacillus buchneri genotyping on the basis of clustered regularly interspaced short palindromic repeat (CRISPR) locus diversity.Appl Environ Microbiol. 2014 Feb;80(3):994-1001. doi: 10.1128/AEM.03015-13. Epub 2013 Nov 22. Appl Environ Microbiol. 2014. PMID: 24271175 Free PMC article.
-
[CRISPR/Cas systems in genome engineering of bacteriophages].Yi Chuan. 2018 May 20;40(5):378-389. doi: 10.16288/j.yczz.17-419. Yi Chuan. 2018. PMID: 29785946 Review. Chinese.
-
Exploiting CRISPR/Cas: interference mechanisms and applications.Int J Mol Sci. 2013 Jul 12;14(7):14518-31. doi: 10.3390/ijms140714518. Int J Mol Sci. 2013. PMID: 23857052 Free PMC article. Review.
Cited by
-
Risk Factors of Clonally Related, Multi, and Extensively Drug-Resistant Acinetobacter baumannii in Severely Ill COVID-19 Patients.Can J Infect Dis Med Microbiol. 2023 Feb 13;2023:3139270. doi: 10.1155/2023/3139270. eCollection 2023. Can J Infect Dis Med Microbiol. 2023. PMID: 36814503 Free PMC article.
-
Broadscale phage therapy is unlikely to select for widespread evolution of bacterial resistance to virus infection.Virus Evol. 2020 Aug 25;6(2):veaa060. doi: 10.1093/ve/veaa060. eCollection 2020 Jul. Virus Evol. 2020. PMID: 33365149 Free PMC article.
-
CRISPR Typing of Clinical Strains of Salmonella spp. Isolated in Tehran, Iran.Iran J Public Health. 2023 Aug;52(8):1758-1763. doi: 10.18502/ijph.v52i8.13415. Iran J Public Health. 2023. PMID: 37744550 Free PMC article.
-
The phylogenomics of CRISPR-Cas system and revelation of its features in Salmonella.Sci Rep. 2020 Dec 3;10(1):21156. doi: 10.1038/s41598-020-77890-6. Sci Rep. 2020. PMID: 33273523 Free PMC article.
-
Virulence Variation of Salmonella Gallinarum Isolates through SpvB by CRISPR Sequence Subtyping, 2014 to 2018.Animals (Basel). 2020 Dec 9;10(12):2346. doi: 10.3390/ani10122346. Animals (Basel). 2020. PMID: 33317043 Free PMC article.
References
-
- Ranjbar R., Karami A., Farshad S., Giammanco G.M., Mammina C. Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. New Microbiol. 2014;37(1):1–15. - PubMed
-
- Pfaller M.A. Molecular epidemiology in the care of patients. Arch. Pathol. Lab. Med. 1999;123(11):1007–1010. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources