Disease-driving CD4+ T cell clonotypes persist for decades in celiac disease
- PMID: 29757191
- PMCID: PMC5983310
- DOI: 10.1172/JCI98819
Disease-driving CD4+ T cell clonotypes persist for decades in celiac disease
Abstract
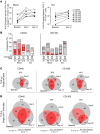
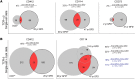
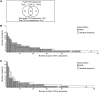
Little is known about the repertoire dynamics and persistence of pathogenic T cells in HLA-associated disorders. In celiac disease, a disorder with a strong association with certain HLA-DQ allotypes, presumed pathogenic T cells can be visualized and isolated with HLA-DQ:gluten tetramers, thereby enabling further characterization. Single and bulk populations of HLA-DQ:gluten tetramer-sorted CD4+ T cells were analyzed by high-throughput DNA sequencing of rearranged TCR-α and -β genes. Blood and gut biopsy samples from 21 celiac disease patients, taken at various stages of disease and in intervals of weeks to decades apart, were examined. Persistence of the same clonotypes was seen in both compartments over decades, with up to 53% overlap between samples obtained 16 to 28 years apart. Further, we observed that the recall response following oral gluten challenge was dominated by preexisting CD4+ T cell clonotypes. Public features were frequent among gluten-specific T cells, as 10% of TCR-α, TCR-β, or paired TCR-αβ amino acid sequences of total 1813 TCRs generated from 17 patients were observed in 2 or more patients. In established celiac disease, the T cell clonotypes that recognize gluten are persistent for decades, making up fixed repertoires that prevalently exhibit public features. These T cells represent an attractive therapeutic target.
Keywords: Autoimmunity; Immunology; T cells; T-cell receptor.
Conflict of interest statement
Figures

Comment in
-
T cells in coeliac disease: a rational target for diagnosis and therapy.Nat Rev Gastroenterol Hepatol. 2018 Oct;15(10):583-584. doi: 10.1038/s41575-018-0043-4. Nat Rev Gastroenterol Hepatol. 2018. PMID: 29973675 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

