Lessons learned from merging wet lab experiments with molecular simulation to improve mAb humanization
- PMID: 29757445
- PMCID: PMC6277173
- DOI: 10.1093/protein/gzy009
Lessons learned from merging wet lab experiments with molecular simulation to improve mAb humanization
Abstract
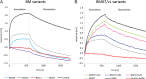
Humanized monoclonal antibodies (mAbs) are among the most promising modern therapeutics, but defined engineering strategies are still not available. Antibody humanization often leads to a loss of affinity, as it is the case for our model antibody Ab2/3H6 (PDB entry 3BQU). Identifying appropriate back-to-mouse mutations is needed to restore binding affinity, but highly challenging. In order to get more insight, we have applied molecular dynamics simulations and correlated them to antibody binding and expression in wet lab experiments. In this study, we discuss six mAb variants and investigate a tyrosine conglomeration, an isopolar substitution and the improvement of antibody binding towards wildtype affinity. In the 3D structure of the mouse wildtype, residue R94h is surrounded by three tyrosines which form a so-called 'tyrosine cage'. We demonstrate that the tyrosine cage has a supporting function for the CDRh3 loop conformation. The isopolar substitution is not able to mimic the function appropriately. Finally, we show that additional light chain mutations can restore binding to wildtype-comparable level, and also improve the expression of the mAb significantly. We conclude that the variable light chain of Ab2/3H6 is of underestimated importance for the interaction with its antigen mAb 2F5.
Figures



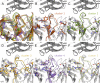
Similar articles
-
Humanization strategies for an anti-idiotypic antibody mimicking HIV-1 gp41.Protein Eng Des Sel. 2010 Dec;23(12):947-54. doi: 10.1093/protein/gzq092. Epub 2010 Oct 30. Protein Eng Des Sel. 2010. PMID: 21037278
-
Antibody humanization by molecular dynamics simulations-in-silico guided selection of critical backmutations.J Mol Recognit. 2016 Jun;29(6):266-75. doi: 10.1002/jmr.2527. Epub 2016 Jan 8. J Mol Recognit. 2016. PMID: 26748949 Free PMC article.
-
Humanization of a phosphothreonine peptide-specific chicken antibody by combinatorial library optimization of the phosphoepitope-binding motif.Biochem Biophys Res Commun. 2015 Jul 31;463(3):414-20. doi: 10.1016/j.bbrc.2015.05.086. Epub 2015 May 30. Biochem Biophys Res Commun. 2015. PMID: 26036575
-
Antibody humanization methods - a review and update.Biotechnol Genet Eng Rev. 2013;29:175-86. doi: 10.1080/02648725.2013.801235. Epub 2013 Aug 2. Biotechnol Genet Eng Rev. 2013. PMID: 24568279 Review.
-
Antibody humanization methods for development of therapeutic applications.Monoclon Antib Immunodiagn Immunother. 2014 Apr;33(2):67-73. doi: 10.1089/mab.2013.0080. Monoclon Antib Immunodiagn Immunother. 2014. PMID: 24746146 Review.
Cited by
-
Pembrolizumab Induces an Unexpected Conformational Change in the CC'-loop of PD-1.Cancers (Basel). 2020 Dec 22;13(1):5. doi: 10.3390/cancers13010005. Cancers (Basel). 2020. PMID: 33375020 Free PMC article.
-
Systematic testing and specificity mapping of alloantigen-specific chimeric antigen receptors in regulatory T cells.JCI Insight. 2019 Mar 21;4(6):e123672. doi: 10.1172/jci.insight.123672. eCollection 2019 Mar 21. JCI Insight. 2019. PMID: 30753169 Free PMC article.
-
A Comprehensive Review of the Mechanisms and Clinical Development of Monoclonal Antibodies in Cancer Therapy.Adv Exp Med Biol. 2025;1479:181-203. doi: 10.1007/5584_2024_838. Adv Exp Med Biol. 2025. PMID: 39666264 Review.
-
Germinality does not necessarily define mAb expression and thermal stability.Appl Microbiol Biotechnol. 2019 Sep;103(18):7505-7518. doi: 10.1007/s00253-019-09998-3. Epub 2019 Jul 26. Appl Microbiol Biotechnol. 2019. PMID: 31350616 Free PMC article.
References
-
- Almagro J.C. (2004) J. Mol. Recognit., 17, 132–143. - PubMed
-
- Bentley K.J., Gewert R. and Harris W.J. (1998) Hybridoma, 17, 559–567. - PubMed
-
- Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A. and Haak J.R. (1984) J. Chem. Phys., 81, 3684–3690.
-
- Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F. and Hermans J. (1981) Pullman B. (ed),. Intermolecular Forces. Reidel, Dordrecht, The Netherlands, pp. 331–342.
-
- Bernett M.J., Karki S., Moore G.L. et al. . (2010) J. Mol. Biol., 396, 1474–1490. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

