Characterization of Rhamnolipids Produced by an Arctic Marine Bacterium from the Pseudomonas fluorescence Group
- PMID: 29758007
- PMCID: PMC5983294
- DOI: 10.3390/md16050163
Characterization of Rhamnolipids Produced by an Arctic Marine Bacterium from the Pseudomonas fluorescence Group
Abstract
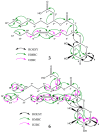
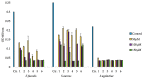
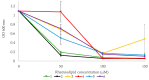
The marine environment is a rich source of biodiversity, including microorganisms that have proven to be prolific producers of bioactive secondary metabolites. Arctic seas are less explored than warmer, more accessible areas, providing a promising starting point to search for novel bioactive compounds. In the present work, an Arctic marine Pseudomonas sp. belonging to the Pseudomonas (P.) fluorescence group was cultivated in four different media in an attempt to activate biosynthetic pathways leading to the production of antibacterial and anticancer compounds. Culture extracts were pre-fractionated and screened for antibacterial and anticancer activities. One fraction from three of the four growth conditions showed inhibitory activity towards bacteria and cancer cells. The active fractions were dereplicated using molecular networking based on MS/MS fragmentation data, indicating the presence of a cluster of related rhamnolipids. Six compounds were isolated using HPLC and mass-guided fractionation, and by interpreting data from NMR and high-resolution MS/MS analysis; the structures of the compounds were determined to be five mono-rhamnolipids and the lipid moiety of one of the rhamnolipids. Molecular networking proved to be a valuable tool for dereplication of these related compounds, and for the first time, five mono-rhamnolipids from a bacterium within the P. fluorescence group were characterized, including one new mono-rhamnolipid.
Keywords: OSMAC (one strain, many compounds); arctic bacteria; bioactive; molecular networking; rhamnolipids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hubert J., Nuzillard J.-M., Renault J.-H. Dereplication strategies in natural product research: How many tools and methodologies behind the same concept? Phytochem. Rev. 2017;16:55–95. doi: 10.1007/s11101-015-9448-7. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

