Genomic evidence that the live Chlamydia abortus vaccine strain 1B is not attenuated and has the potential to cause disease
- PMID: 29759381
- PMCID: PMC6005232
- DOI: 10.1016/j.vaccine.2018.05.042
Genomic evidence that the live Chlamydia abortus vaccine strain 1B is not attenuated and has the potential to cause disease
Abstract
Background: The live, temperature-attenuated vaccine strain 1B of Chlamydia abortus, the aetiological agent of ovine enzootic abortion (OEA), has been implicated in cases of vaccine breakdown. The aim of this study was to understand the nature of this attenuation through sequencing of the vaccine parent strain (AB7) and the derived mutant strains 1B and 1H, as well as to clarify the role of the vaccine strain in causing disease through comparative whole genome analysis.
Methods: Whole genome sequencing was performed on: vaccine parent strain AB7; N-methyl-N'-nitro-N-nitrosoguanidine (NTG)-induced temperature attenuated mutant strain 1B grown from the commercial live vaccines Cevac Chlamydia and Enzovax; strain 1H a reverted NTG mutant; and 5 strains isolated from cases of OEA originating from animals from the original vaccine safety trial (2 strains) or from vaccinated ewes or ewes exposed to vaccinated animals (3 strains).
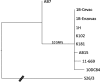
Results: We confirmed that AB7 is in a different lineage from the reference strain S26/3. The genome of vaccine strain 1B contains ten single nucleotide polymorphisms (SNPs) created by the NTG treatment, which are identical to those found in strain 1H. The strains from OEA cases also cluster phylogenetically very tightly with these vaccine strains.
Conclusions: The results show that C. abortus vaccine strain 1B has an identical genome sequence to the non-attenuated "reverted mutant" strain 1H. Thus, the protection of the 1B vaccine is unlikely to be due to the NTG induced SNPs and is more likely caused by the administration of high doses of C. abortus elementary bodies that stimulate protective immunity. Vaccine-identical strains were also isolated from cases of disease, as well as strains which had acquired 1-3 SNPs, including an animal that had not been vaccinated with either of the commercial live OEA vaccines, indicating that the 1B vaccine strain may be circulating and causing disease.
Keywords: Chlamydia abortus; Genome analysis; Ovine enzootic abortion; Single nucleotide polymorphisms; Vaccination.
Copyright © 2018. Published by Elsevier Ltd.
Figures

References
-
- Longbottom D., Coulter L.J. Animal chlamydioses and zoonotic implications. J Comp Pathol. 2003;128:217–244. - PubMed
-
- Aitken I.D., Longbottom D. Chlamydial abortion. In: Aitken I.D., editor. Diseases of sheep. 4th ed. Blackwell Publishing; Oxford: 2007. pp. 105–111.
-
- Longbottom D., Livingstone M. Vaccination against chlamydial infections of man and animals. Vet J. 2006;171:263–275. - PubMed
-
- Faye P.A., Charton A., Mage C., Bernard C., Le Layec C. Proprietés hémagglutinantes du virus de l’avortement enzootique des petits ruminants (souches de Rakeia d’origine ovine et caprine) Bull Acad Vet Fr. 1972;45:169–173.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

