Size-tagged preferred ends in maternal plasma DNA shed light on the production mechanism and show utility in noninvasive prenatal testing
- PMID: 29760053
- PMCID: PMC5984542
- DOI: 10.1073/pnas.1804134115
Size-tagged preferred ends in maternal plasma DNA shed light on the production mechanism and show utility in noninvasive prenatal testing
Abstract
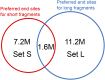
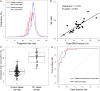
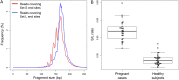
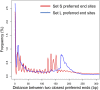
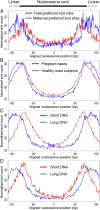
Cell-free DNA in human plasma is nonrandomly fragmented and reflects genomewide nucleosomal organization. Previous studies had demonstrated tissue-specific preferred end sites in plasma DNA of pregnant women. In this study, we performed integrative analysis of preferred end sites with the size characteristics of plasma DNA fragments. We mined the preferred end sites in short and long plasma DNA molecules separately and found that these "size-tagged" ends showed improved accuracy in fetal DNA fraction estimation and enhanced noninvasive fetal trisomy 21 testing. Further analysis revealed that the fetal and maternal preferred ends were generated from different locations within the nucleosomal structure. Hence, fetal DNA was frequently cut within the nucleosome core while maternal DNA was mostly cut within the linker region. We further demonstrated that the nucleosome accessibility in placental cells was higher than that for white blood cells, which might explain the difference in the cutting positions and the shortness of fetal DNA in maternal plasma. Interestingly, short and long size-tagged ends were also observable in the plasma of nonpregnant healthy subjects and demonstrated size differences similar to those in the pregnant samples. Because the nonpregnant samples did not contain fetal DNA, the data suggested that the interrelationship of preferred DNA ends, chromatin accessibility, and plasma DNA size profile is likely a general one, extending beyond the context of pregnancy. Plasma DNA fragment end patterns have thus shed light on production mechanisms and show utility in future developments in plasma DNA-based noninvasive molecular diagnostics.
Keywords: circulating cell-free DNA; liquid biopsy; nucleosome structure; prenatal diagnosis; size-based molecular diagnostics.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: K.C.A.C., R.W.K.C., and Y.M.D.L. hold equities in DRA and Grail, are consultants to Grail, and receive research funding from Grail/Cirina. P.J. is a consultant to Xcelom and Grail. Y.M.D.L. is a scientific cofounder and a member of the scientific advisory board for Grail.
Figures



References
-
- Lo YMD, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. - PubMed
-
- Lo YMD, et al. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet. 1998;351:1329–1330. - PubMed
-
- Stroun M, et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46:318–322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

