Pseudomonas aeruginosa transcriptome during human infection
- PMID: 29760087
- PMCID: PMC5984494
- DOI: 10.1073/pnas.1717525115
Pseudomonas aeruginosa transcriptome during human infection
Abstract
Laboratory experiments have uncovered many basic aspects of bacterial physiology and behavior. After the past century of mostly in vitro experiments, we now have detailed knowledge of bacterial behavior in standard laboratory conditions, but only a superficial understanding of bacterial functions and behaviors during human infection. It is well-known that the growth and behavior of bacteria are largely dictated by their environment, but how bacterial physiology differs in laboratory models compared with human infections is not known. To address this question, we compared the transcriptome of Pseudomonas aeruginosa during human infection to that of P. aeruginosa in a variety of laboratory conditions. Several pathways, including the bacterium's primary quorum sensing system, had significantly lower expression in human infections than in many laboratory conditions. On the other hand, multiple genes known to confer antibiotic resistance had substantially higher expression in human infection than in laboratory conditions, potentially explaining why antibiotic resistance assays in the clinical laboratory frequently underestimate resistance in patients. Using a standard machine learning technique known as support vector machines, we identified a set of genes whose expression reliably distinguished in vitro conditions from human infections. Finally, we used these support vector machines with binary classification to force P. aeruginosa mouse infection transcriptomes to be classified as human or in vitro. Determining what differentiates our current models from clinical infections is important to better understand bacterial infections and will be necessary to create model systems that more accurately capture the biology of infection.
Keywords: Pseudomonas aeruginosa; chronic wounds; cystic fibrosis; human transcriptome; machine learning.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
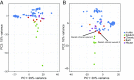
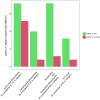
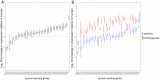
Comment in
-
Direct In Vivo Microbial Transcriptomics During Infection.Trends Microbiol. 2018 Sep;26(9):732-735. doi: 10.1016/j.tim.2018.07.002. Epub 2018 Jul 31. Trends Microbiol. 2018. PMID: 30075901
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

