Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility
- PMID: 29760102
- PMCID: PMC5984533
- DOI: 10.1073/pnas.1802879115
Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility
Abstract
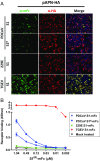
Porcine deltacoronavirus (PDCoV), identified in 2012, is a common enteropathogen of swine with worldwide distribution. The source and evolutionary history of this virus is, however, unknown. PDCoV belongs to the Deltacoronavirus genus that comprises predominantly avian CoV. Phylogenetic analysis suggests that PDCoV originated relatively recently from a host-switching event between birds and mammals. Insight into receptor engagement by PDCoV may shed light into such an exceptional phenomenon. Here we report that PDCoV employs host aminopeptidase N (APN) as an entry receptor and interacts with APN via domain B of its spike (S) protein. Infection of porcine cells with PDCoV was drastically reduced by APN knockout and rescued after reconstitution of APN expression. In addition, we observed that PDCoV efficiently infects cells of unusual broad species range, including human and chicken. Accordingly, PDCoV S was found to target the phylogenetically conserved catalytic domain of APN. Moreover, transient expression of porcine, feline, human, and chicken APN renders cells susceptible to PDCoV infection. Binding of PDCoV to an interspecies conserved site on APN may facilitate direct transmission of PDCoV to nonreservoir species, including humans, potentially reflecting the mechanism that enabled a virus, ancestral to PDCoV, to breach the species barrier between birds and mammals. The APN cell surface protein is also used by several members of the Alphacoronavirus genus. Hence, our data constitute the second identification of CoVs from different genera that use the same receptor, implying that CoV receptor selection is subjected to specific restrictions that are still poorly understood.
Keywords: APN; PDCoV; cross-species transmission; receptor; spike.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- World Health Organization 2015 Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. Available at www.who.int/csr/sars/country/table2004_04_21/en/. Accessed January 29, 2018.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

