Metabolic regulation of chromatin modifications and gene expression
- PMID: 29760106
- PMCID: PMC6028552
- DOI: 10.1083/jcb.201803061
Metabolic regulation of chromatin modifications and gene expression
Abstract
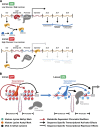
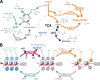
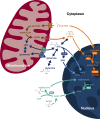
Dynamic regulation of gene expression in response to changing local conditions is critical for the survival of all organisms. In metazoans, coherent regulation of gene expression programs underlies the development of functionally distinct cell lineages. The cooperation between transcription factors and the chromatin landscape enables precise control of gene expression in response to cell-intrinsic and cell-extrinsic signals. Many of the chemical modifications that decorate DNA and histones are adducts derived from intermediates of cellular metabolic pathways. In addition, several of the enzymes that can remove these marks use metabolites as part of their enzymatic reaction. These observations have led to the hypothesis that fluctuations in metabolite levels influence the deposition and removal of chromatin modifications. In this review, we consider the emerging evidence that cellular metabolic activity contributes to gene expression and cell fate decisions through metabolite-dependent effects on chromatin organization.
© 2018 Schvartzman et al.
Figures
References
-
- Baysal B.E., Ferrell R.E., Willett-Brozick J.E., Lawrence E.C., Myssiorek D., Bosch A., van der Mey A., Taschner P.E., Rubinstein W.S., Myers E.N., et al. 2000. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 287:848–851. 10.1126/science.287.5454.848 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

