Developmental Sensitivity in Schistosoma mansoni to Puromycin To Establish Drug Selection of Transgenic Schistosomes
- PMID: 29760143
- PMCID: PMC6105839
- DOI: 10.1128/AAC.02568-17
Developmental Sensitivity in Schistosoma mansoni to Puromycin To Establish Drug Selection of Transgenic Schistosomes
Abstract
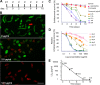
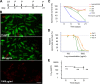
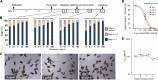
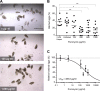
Schistosomiasis is considered the most important disease caused by helminth parasites, in terms of morbidity and mortality. Tools to facilitate gain- and loss-of-function approaches can be expected to precipitate the discovery of novel interventions, and drug selection of transgenic schistosomes would facilitate the establishment of stable lines of engineered parasites. Sensitivity of developmental stages of schistosomes to the aminonucleoside antibiotic puromycin was investigated. For the schistosomulum and sporocyst stages, viability was quantified by fluorescence microscopy following dual staining with fluorescein diacetate and propidium iodine. By 6 days in culture, the 50% lethal concentration (LC50) for schistosomula was 19 μg/ml whereas the sporocysts were 45-fold more resilient. Puromycin potently inhibited the development of in vitro-laid eggs (LC50, 68 ng/ml) but was less effective against liver eggs (LC50, 387 μg/ml). Toxicity for adult stages was evaluated using the xCELLigence-based, real-time motility assay (xWORM), which revealed LC50s after 48 h of 4.9 and 17.3 μg/ml for male and female schistosomes, respectively. Also, schistosomula transduced with pseudotyped retrovirus encoding the puromycin resistance marker were partially rescued when cultured in the presence of the antibiotic. Together, these findings will facilitate selection on puromycin of transgenic schistosomes and the enrichment of cultures of transgenic eggs and sporocysts to facilitate the establishment of schistosome transgenic lines. Streamlining schistosome transgenesis with drug selection will open new avenues to understand parasite biology and hopefully lead to new interventions for this neglected tropical disease.
Keywords: antibiotic susceptibility; drug selection; functional genomics; puromycin; schistosomiasis.
Copyright © 2018 American Society for Microbiology.
Figures


References
-
- Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, Aslett MA, Bartholomeu DC, Blandin G, Caffrey CR, Coghlan A, Coulson R, Day TA, Delcher A, DeMarco R, Djikeng A, Eyre T, Gamble JA, Ghedin E, Gu Y, Hertz-Fowler C, Hirai H, Hirai Y, Houston R, Ivens A, Johnston DA, Lacerda D, Macedo CD, McVeigh P, Ning Z, Oliveira G, Overington JP, Parkhill J, Pertea M, Pierce RJ, Protasio AV, Quail MA, Rajandream MA, Rogers J, Sajid M, Salzberg SL, Stanke M, Tivey AR, White O, Williams DL, Wortman J, Wu W, Zamanian M, Zerlotini A, Fraser-Liggett CM, Barrell BG, El-Sayed NM. 2009. The genome of the blood fluke Schistosoma mansoni. Nature 460:352–358. doi: 10.1038/nature08160. - DOI - PMC - PubMed
-
- Protasio AV, Tsai IJ, Babbage A, Nichol S, Hunt M, Aslett MA, De Silva N, Velarde GS, Anderson TJ, Clark RC, Davidson C, Dillon GP, Holroyd NE, LoVerde PT, Lloyd C, McQuillan J, Oliveira G, Otto TD, Parker-Manuel SJ, Quail MA, Wilson RA, Zerlotini A, Dunne DW, Berriman M. 2012. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl Trop Dis 6:e1455. doi: 10.1371/journal.pntd.0001455. - DOI - PMC - PubMed
-
- Young ND, Jex AR, Li B, Liu S, Yang L, Xiong Z, Li Y, Cantacessi C, Hall RS, Xu X, Chen F, Wu X, Zerlotini A, Oliveira G, Hofmann A, Zhang G, Fang X, Kang Y, Campbell BE, Loukas A, Ranganathan S, Rollinson D, Rinaldi G, Brindley PJ, Yang H, Wang J, Gasser RB. 2012. Whole-genome sequence of Schistosoma haematobium. Nat Genet 44:221–225. doi: 10.1038/ng.1065. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

