Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus
- PMID: 29760169
- PMCID: PMC6109274
- DOI: 10.1136/gutjnl-2018-315988
Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus
Abstract
Objective: The initial colonisation of the human microbiota and the impact of maternal health on neonatal microbiota at birth remain largely unknown. The aim of our study is to investigate the possible dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus (GDM) and to estimate the potential risks of the microbial shift to neonates.
Design: Pregnant women and neonates suffering from GDM were enrolled and 581 maternal (oral, intestinal and vaginal) and 248 neonatal (oral, pharyngeal, meconium and amniotic fluid) samples were collected. To avoid vaginal bacteria contaminations, the included neonates were predominantly delivered by C-section, with their samples collected within seconds of delivery.
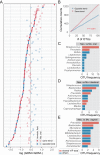
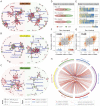
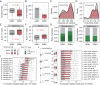
Results: Numerous and diverse bacterial taxa were identified from the neonatal samples, and the samples from different neonatal body sites were grouped into distinct clusters. The microbiota of pregnant women and neonates was remarkably altered in GDM, with a strong correlation between certain discriminatory bacteria and the oral glucose tolerance test. Microbes varying by the same trend across the maternal and neonatal microbiota were observed, revealing the intergenerational concordance of microbial variation associated with GDM. Furthermore, lower evenness but more depletion of KEGG orthologues and higher abundance of some viruses (eg, herpesvirus and mastadenovirus) were observed in the meconium microbiota of neonates associated with GDM.
Conclusion: GDM can alter the microbiota of both pregnant women and neonates at birth, which sheds light on another form of inheritance and highlights the importance of understanding the formation of early-life microbiome.
Keywords: GDM; microbiota; neonate; pregnancy.
© Article author(s) (or their employer(s) unless otherwise stated in the text of the article) 2018. All rights reserved. No commercial use is permitted unless otherwise expressly granted.
Conflict of interest statement
Competing interests: None declared.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
