Neuron-Specific HuR-Deficient Mice Spontaneously Develop Motor Neuron Disease
- PMID: 29760195
- PMCID: PMC6008238
- DOI: 10.4049/jimmunol.1701501
Neuron-Specific HuR-Deficient Mice Spontaneously Develop Motor Neuron Disease
Erratum in
-
Correction: Neuron-Specific HuR-Deficient Mice Spontaneously Develop Motor Neuron Disease.J Immunol. 2018 Sep 1;201(5):1600. doi: 10.4049/jimmunol.1800930. Epub 2018 Jul 23. J Immunol. 2018. PMID: 30037850 No abstract available.
Abstract
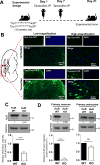
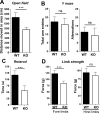
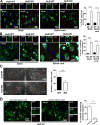
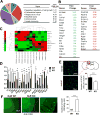
Human Ag R (HuR) is an RNA binding protein in the ELAVL protein family. To study the neuron-specific function of HuR, we generated inducible, neuron-specific HuR-deficient mice of both sexes. After tamoxifen-induced deletion of HuR, these mice developed a phenotype consisting of poor balance, decreased movement, and decreased strength. They performed significantly worse on the rotarod test compared with littermate control mice, indicating coordination deficiency. Using the grip-strength test, it was also determined that the forelimbs of neuron-specific HuR-deficient mice were much weaker than littermate control mice. Immunostaining of the brain and cervical spinal cord showed that HuR-deficient neurons had increased levels of cleaved caspase-3, a hallmark of cell apoptosis. Caspase-3 cleavage was especially strong in pyramidal neurons and α motor neurons of HuR-deficient mice. Genome-wide microarray and real-time PCR analysis further indicated that HuR deficiency in neurons resulted in altered expression of genes in the brain involved in cell growth, including trichoplein keratin filament-binding protein, Cdkn2c, G-protein signaling modulator 2, immediate early response 2, superoxide dismutase 1, and Bcl2. The additional enriched Gene Ontology terms in the brain tissues of neuron-specific HuR-deficient mice were largely related to inflammation, including IFN-induced genes and complement components. Importantly, some of these HuR-regulated genes were also significantly altered in the brain and spinal cord of patients with amyotrophic lateral sclerosis. Additionally, neuronal HuR deficiency resulted in the redistribution of TDP43 to cytosolic granules, which has been linked to motor neuron disease. Taken together, we propose that this neuron-specific HuR-deficient mouse strain can potentially be used as a motor neuron disease model.
Copyright © 2018 by The American Association of Immunologists, Inc.
Figures

Similar articles
-
Neuroprotective effects of the Sigma-1 receptor (S1R) agonist PRE-084, in a mouse model of motor neuron disease not linked to SOD1 mutation.Neurobiol Dis. 2014 Feb;62:218-32. doi: 10.1016/j.nbd.2013.10.010. Epub 2013 Oct 16. Neurobiol Dis. 2014. PMID: 24141020
-
The Overexpression of TDP-43 Protein in the Neuron and Oligodendrocyte Cells Causes the Progressive Motor Neuron Degeneration in the SOD1 G93A Transgenic Mouse Model of Amyotrophic Lateral Sclerosis.Int J Biol Sci. 2016 Aug 15;12(9):1140-9. doi: 10.7150/ijbs.15938. eCollection 2016. Int J Biol Sci. 2016. PMID: 27570488 Free PMC article.
-
An early onset progressive motor neuron disorder in Scyl1-deficient mice is associated with mislocalization of TDP-43.J Neurosci. 2012 Nov 21;32(47):16560-73. doi: 10.1523/JNEUROSCI.1787-12.2012. J Neurosci. 2012. PMID: 23175812 Free PMC article.
-
Role of complement in motor neuron disease: animal models and therapeutic potential of complement inhibitors.Adv Exp Med Biol. 2008;632:143-58. Adv Exp Med Biol. 2008. PMID: 19025120 Review.
-
Motor neuron disease in vitro: the use of cultured motor neurons to study amyotrophic lateral sclerosis.Eur J Pharmacol. 2000 Sep 29;405(1-3):285-95. doi: 10.1016/s0014-2999(00)00560-4. Eur J Pharmacol. 2000. PMID: 11033335 Review.
Cited by
-
Effects of genetic ablation and pharmacological inhibition of HuR on gene expression, iron metabolism, and hormone levels.BMC Biol. 2025 Jan 23;23(1):24. doi: 10.1186/s12915-025-02131-z. BMC Biol. 2025. PMID: 39849491 Free PMC article.
-
Unraveling the Pathways to Neuronal Homeostasis and Disease: Mechanistic Insights into the Role of RNA-Binding Proteins and Associated Factors.Int J Mol Sci. 2018 Aug 3;19(8):2280. doi: 10.3390/ijms19082280. Int J Mol Sci. 2018. PMID: 30081499 Free PMC article. Review.
-
Tissue-specific mechanisms of alternative polyadenylation: Testis, brain, and beyond (2018 update).Wiley Interdiscip Rev RNA. 2019 Jul;10(4):e1526. doi: 10.1002/wrna.1526. Epub 2019 Feb 27. Wiley Interdiscip Rev RNA. 2019. PMID: 30816016 Free PMC article. Review.
-
ELAV/Hu RNA-binding protein family: key regulators in neurological disorders, cancer, and other diseases.RNA Biol. 2025 Dec;22(1):1-11. doi: 10.1080/15476286.2025.2471133. Epub 2025 Mar 20. RNA Biol. 2025. PMID: 40000387 Free PMC article. Review.
-
Colocalization of TDP-43 and stress granules at the early stage of TDP-43 aggregation in amyotrophic lateral sclerosis.Brain Pathol. 2024 Mar;34(2):e13215. doi: 10.1111/bpa.13215. Epub 2023 Oct 4. Brain Pathol. 2024. PMID: 37793650 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

