GLUCOSAMINE INOSITOLPHOSPHORYLCERAMIDE TRANSFERASE1 (GINT1) Is a GlcNAc-Containing Glycosylinositol Phosphorylceramide Glycosyltransferase
- PMID: 29760197
- PMCID: PMC6053017
- DOI: 10.1104/pp.18.00396
GLUCOSAMINE INOSITOLPHOSPHORYLCERAMIDE TRANSFERASE1 (GINT1) Is a GlcNAc-Containing Glycosylinositol Phosphorylceramide Glycosyltransferase
Abstract
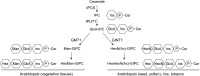
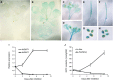
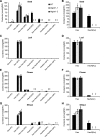
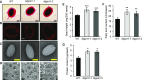
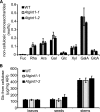
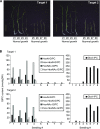
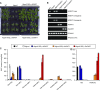
Glycosylinositol phosphorylceramides (GIPCs), which have a ceramide core linked to a glycan headgroup of varying structures, are the major sphingolipids in the plant plasma membrane. Recently, we identified the major biosynthetic genes for GIPC glycosylation in Arabidopsis (Arabidopsis thaliana) and demonstrated that the glycan headgroup is essential for plant viability. However, the function of GIPCs and the significance of their structural variation are poorly understood. Here, we characterized the Arabidopsis glycosyltransferase GLUCOSAMINE INOSITOLPHOSPHORYLCERAMIDE TRANSFERASE1 (GINT1) and showed that it is responsible for the glycosylation of a subgroup of GIPCs found in seeds and pollen that contain GlcNAc and GlcN [collectively GlcN(Ac)]. In Arabidopsis gint1 plants, loss of the GlcN(Ac) GIPCs did not affect vegetative growth, although seed germination was less sensitive to abiotic stress than in wild-type plants. However, in rice, where GlcN(Ac) containing GIPCs are the major GIPC subgroup in vegetative tissue, loss of GINT1 was seedling lethal. Furthermore, we could produce, de novo, "rice-like" GlcN(Ac) GIPCs in Arabidopsis leaves, which allowed us to test the function of different sugars in the GIPC headgroup. This study describes a monocot GIPC biosynthetic enzyme and shows that its Arabidopsis homolog has the same biochemical function. We also identify a possible role for GIPCs in maintaining cell-cell adhesion.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures



References
-
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 - PubMed
-
- Buré C, Cacas JL, Wang F, Gaudin K, Domergue F, Mongrand S, Schmitter JM (2011) Fast screening of highly glycosylated plant sphingolipids by tandem mass spectrometry. Rapid Commun Mass Spectrom 25: 3131–3145 - PubMed
-
- Busse-Wicher M, Wicher KB, Kusche-Gullberg M (2014) The exostosin family: proteins with many functions. Matrix Biol 35: 25–33 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

