Updates to Clostridium difficile Spore Germination
- PMID: 29760211
- PMCID: PMC6060349
- DOI: 10.1128/JB.00218-18
Updates to Clostridium difficile Spore Germination
Abstract
Germination of Clostridium difficile spores is a crucial early requirement for colonization of the gastrointestinal tract. Likewise, C. difficile cannot cause disease pathologies unless its spores germinate into metabolically active, toxin-producing cells. Recent advances in our understanding of C. difficile spore germination mechanisms indicate that this process is both complex and unique. This review defines unique aspects of the germination pathways of C. difficile and compares them to those of two other well-studied organisms, Bacillus anthracis and Clostridium perfringensC. difficile germination is unique, as C. difficile does not contain any orthologs of the traditional GerA-type germinant receptor complexes and is the only known sporeformer to require bile salts in order to germinate. While recent advances describing C. difficile germination mechanisms have been made on several fronts, major gaps in our understanding of C. difficile germination signaling remain. This review provides an updated, in-depth summary of advances in understanding of C. difficile germination and potential avenues for the development of therapeutics, and discusses the major discrepancies between current models of germination and areas of ongoing investigation.
Keywords: Clostridium difficile; germination; spores.
Copyright © 2018 American Society for Microbiology.
Figures
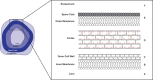



References
-
- Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi:10.1056/NEJMoa1408913. - DOI - PMC - PubMed
-
- Davies KA, Ashwin H, Longshaw CM, Burns DA, Davis GL, Wilcox MH, EUCLID Study Group. 2016. Diversity of Clostridium difficile PCR ribotypes in Europe: results from the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID), 2012 and 2013. Euro Surveill 21(29):pii=30294. doi:10.2807/1560-7917.ES.2016.21.29.30294. - DOI - PubMed
-
- Kuijper EJ, Coignard B, Tull P, ESCMID Study Group for Clostridium difficile, EU Member States, European Centre for Disease Prevention and Control . 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect 12(Suppl 6):S2–S18. doi:10.1111/j.1469-0691.2006.01580.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

