Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus
- PMID: 29760414
- PMCID: PMC6015761
- DOI: 10.1038/s41557-018-0039-2
Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus
Abstract
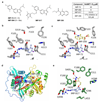
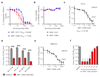
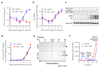
Rhinoviruses (RVs) are the pathogens most often responsible for the common cold, and are a frequent cause of exacerbations in asthma, chronic obstructive pulmonary disease and cystic fibrosis. Here we report the discovery of IMP-1088, a picomolar dual inhibitor of the human N-myristoyltransferases NMT1 and NMT2, and use it to demonstrate that pharmacological inhibition of host-cell N-myristoylation rapidly and completely prevents rhinoviral replication without inducing cytotoxicity. The identification of cooperative binding between weak-binding fragments led to rapid inhibitor optimization through fragment reconstruction, structure-guided fragment linking and conformational control over linker geometry. We show that inhibition of the co-translational myristoylation of a specific virus-encoded protein (VP0) by IMP-1088 potently blocks a key step in viral capsid assembly, to deliver a low nanomolar antiviral activity against multiple RV strains, poliovirus and foot and-mouth disease virus, and protection of cells against virus-induced killing, highlighting the potential of host myristoylation as a drug target in picornaviral infections.
Conflict of interest statement
A.B., E.W.T., R.J.L., J.A.H. and J.A.B. are inventors on a patent application describing NMT inhibitors including IMP-1031 and IMP-1088 (Bell, A.S.; Tate, E.W.; Leatherbarrow, R.J.; Hutton, J.A.; Brannigan, J.A., “Compounds and their use as inhibitors of
Figures


References
-
- Ritchie AI, et al. Pathogenesis of Viral Infection in Exacerbations of Airway Disease. Ann Am Thorac Soc. 2015;12(Suppl 2):S115–132. - PubMed
-
- Kieninger E, et al. High rhinovirus burden in lower airways of children with cystic fibrosis. Chest. 2013;143:782–790. - PubMed
-
- Flight WG, et al. Incidence and clinical impact of respiratory viruses in adults with cystic fibrosis. Thorax. 2014;69:247–253. - PubMed
-
- Thibaut HJ, De Palma AM, Neyts J. Combating enterovirus replication: state-of-the-art on antiviral research. Biochem Pharmacol. 2012;83:185–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

