Characterization of Wnt and Notch-Responsive Lgr5+ Hair Cell Progenitors in the Striolar Region of the Neonatal Mouse Utricle
- PMID: 29760650
- PMCID: PMC5937014
- DOI: 10.3389/fnmol.2018.00137
Characterization of Wnt and Notch-Responsive Lgr5+ Hair Cell Progenitors in the Striolar Region of the Neonatal Mouse Utricle
Abstract
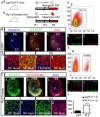
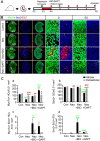
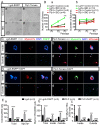
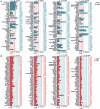
Dysfunctions in hearing and balance are largely connected with hair cell (HC) loss. Although regeneration of HCs in the adult cochlea does not occur, there is still limited capacity for HC regeneration in the mammalian utricle from a distinct population of supporting cells (SCs). In response to HC damage, these Lgr5+ SCs, especially those in the striolar region, can regenerate HCs. In this study, we isolated Lgr5+ SCs and Plp1+ SCs (which originate from the striolar and extrastriolar regions, respectively) from transgenic mice by flow cytometry so as to compare the properties of these two subsets of SCs. We found that the Lgr5+ progenitors had greater proliferation and HC regeneration ability than the Plp1+ SCs and that the Lgr5+ progenitors responded more strongly to Wnt and Notch signaling than Plp1+ SCs. We then compared the gene expression profiles of the two populations by RNA-Seq and identified several genes that were significantly differentially expressed between the two populations, including genes involved in the cell cycle, transcription and cell signaling pathways. Targeting these genes and pathways might be a potential way to activate HC regeneration.
Keywords: hearing and balance; regeneration; stem cell; supporting cells; utricle.
Figures

Similar articles
-
Characterization of the Transcriptomes of Lgr5+ Hair Cell Progenitors and Lgr5- Supporting Cells in the Mouse Cochlea.Front Mol Neurosci. 2017 Apr 26;10:122. doi: 10.3389/fnmol.2017.00122. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28491023 Free PMC article.
-
Characterization of Lgr5+ Progenitor Cell Transcriptomes after Neomycin Injury in the Neonatal Mouse Cochlea.Front Mol Neurosci. 2017 Jul 4;10:213. doi: 10.3389/fnmol.2017.00213. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28725177 Free PMC article.
-
Lgr5+ cells regenerate hair cells via proliferation and direct transdifferentiation in damaged neonatal mouse utricle.Nat Commun. 2015 Apr 7;6:6613. doi: 10.1038/ncomms7613. Nat Commun. 2015. PMID: 25849379 Free PMC article.
-
Role of Wnt and Notch signaling in regulating hair cell regeneration in the cochlea.Front Med. 2016 Sep;10(3):237-49. doi: 10.1007/s11684-016-0464-9. Epub 2016 Sep 7. Front Med. 2016. PMID: 27527363 Review.
-
Therapeutic Potential of Wnt and Notch Signaling and Epigenetic Regulation in Mammalian Sensory Hair Cell Regeneration.Mol Ther. 2019 May 8;27(5):904-911. doi: 10.1016/j.ymthe.2019.03.017. Epub 2019 Mar 30. Mol Ther. 2019. PMID: 30982678 Free PMC article. Review.
Cited by
-
Hyperoside Attenuate Inflammation in HT22 Cells via Upregulating SIRT1 to Activities Wnt/β-Catenin and Sonic Hedgehog Pathways.Neural Plast. 2021 Jun 10;2021:8706400. doi: 10.1155/2021/8706400. eCollection 2021. Neural Plast. 2021. PMID: 34221003 Free PMC article.
-
Conditional Overexpression of Serpine2 Promotes Hair Cell Regeneration from Lgr5+ Progenitors in the Neonatal Mouse Cochlea.Adv Sci (Weinh). 2025 May;12(18):e2412653. doi: 10.1002/advs.202412653. Epub 2025 Mar 17. Adv Sci (Weinh). 2025. PMID: 40091489 Free PMC article.
-
Co-transduction of dual-adeno-associated virus vectors in the neonatal and adult mouse utricles.Front Mol Neurosci. 2022 Oct 19;15:1020803. doi: 10.3389/fnmol.2022.1020803. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36340687 Free PMC article.
-
Transcriptomic Analysis Reveals an Altered Hcy Metabolism in the Stria Vascularis of the Pendred Syndrome Mouse Model.Neural Plast. 2021 Apr 17;2021:5585394. doi: 10.1155/2021/5585394. eCollection 2021. Neural Plast. 2021. PMID: 33959158 Free PMC article.
-
MECOM promotes supporting cell proliferation and differentiation in cochlea.J Otol. 2022 Apr;17(2):59-66. doi: 10.1016/j.joto.2021.11.002. Epub 2021 Nov 24. J Otol. 2022. PMID: 35949554 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

