Plasma proteomic analysis of systemic lupus erythematosus patients using liquid chromatography/tandem mass spectrometry with label-free quantification
- PMID: 29761050
- PMCID: PMC5947061
- DOI: 10.7717/peerj.4730
Plasma proteomic analysis of systemic lupus erythematosus patients using liquid chromatography/tandem mass spectrometry with label-free quantification
Abstract
Context: Systemic lupus erythematosus (SLE) is a chronic inflammatory autoimmune disease with unknown etiology.
Objective: Human plasma is comprised of over 10 orders of magnitude concentration of proteins and tissue leakages. The changes in the abundance of these proteins have played an important role in various human diseases. Therefore, the research objective of this study is to identify the significantly altered expression levels of plasma proteins from SLE patients compared with healthy controls using proteomic analysis. The plasma proteome profiles of both SLE patients and controls were compared.
Methods: A total of 19 active SLE patients and 12 healthy controls plasma samples were analyzed using high-resolution electrospray ionization liquid chromatography-tandem mass spectrometry (LC-ESI-MS/MS) followed by label-free quantification.
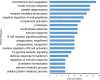
Results: A total of 19 proteins showed a significant level of expression in the comparative LC-ESI-MS/MS triplicate analysis; among these, 14 proteins had >1.5- to three-fold up-regulation and five had <0.2- to 0.6-fold down-regulation. Gene ontology and DAVID (Database Annotation Visualization, and Integrated Discovery) functional enrichment analysis revealed that these proteins are involved in several important biological processes including acute phase inflammatory responses, complement activation, hemostasis, and immune system regulation.
Conclusion: Our study identified a group of differentially expressed proteins in the plasma of SLE patients that are involved in the imbalance of the immune system and inflammatory responses. Therefore, these findings may have the potential to be used as prognostic/diagnostic markers for SLE disease assessment or disease monitoring.
Keywords: Biomarkers; LC-MS/MS; Label-free quantification; Lupus; Plasma; Protein profiling; Proteomic analysis; Proteomics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Abe H, Tsuboi N, Suzuki S, Sakuraba H. Anti-apolipoprotein A-I autoantibody: characterization of monoclonal autoantibodies from patients with systemic lupus erythematosus. Rheumatology. 2001;28:990–995. - PubMed
-
- Aggarwal A, Gupta R, Negi VS, Rajasekhar L, Misra R, Singh P, Chaturvedi V, Sinha S. Urinary haptoglobin, alpha-1 anti-chymotrypsin and retinol binding protein identified by proteomics as potential biomarkers for lupus nephritis. Clinical & Experimental Immunology. 2017;188(2):254–262. doi: 10.1111/cei.12930. - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

