Genomic analysis identifies masqueraders of full-term cerebral palsy
- PMID: 29761117
- PMCID: PMC5945967
- DOI: 10.1002/acn3.551
Genomic analysis identifies masqueraders of full-term cerebral palsy
Abstract
Objective: Cerebral palsy is a common, heterogeneous neurodevelopmental disorder that causes movement and postural disabilities. Recent studies have suggested genetic diseases can be misdiagnosed as cerebral palsy. We hypothesized that two simple criteria, that is, full-term births and nonspecific brain MRI findings, are keys to extracting masqueraders among cerebral palsy cases due to the following: (1) preterm infants are susceptible to multiple environmental factors and therefore demonstrate an increased risk of cerebral palsy and (2) brain MRI assessment is essential for excluding environmental causes and other particular disorders.
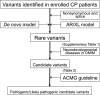
Methods: A total of 107 patients-all full-term births-without specific findings on brain MRI were identified among 897 patients diagnosed with cerebral palsy who were followed at our center. DNA samples were available for 17 of the 107 cases for trio whole-exome sequencing and array comparative genomic hybridization. We prioritized variants in genes known to be relevant in neurodevelopmental diseases and evaluated their pathogenicity according to the American College of Medical Genetics guidelines.
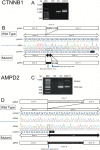
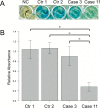
Results: Pathogenic/likely pathogenic candidate variants were identified in 9 of 17 cases (52.9%) within eight genes: CTNNB1,CYP2U1,SPAST,GNAO1,CACNA1A,AMPD2,STXBP1, and SCN2A. Five identified variants had previously been reported. No pathogenic copy number variations were identified. The AMPD2 missense variant and the splice-site variants in CTNNB1 and AMPD2 were validated by in vitro functional experiments.
Interpretation: The high rate of detecting causative genetic variants (52.9%) suggests that patients diagnosed with cerebral palsy in full-term births without specific MRI findings may include genetic diseases masquerading as cerebral palsy.
Figures


References
-
- Rosenbaum P, Paneth N, Leviton A, et al. A report: the definition and classification of cerebral palsy April 2006. Dev Med Child Neurol Suppl 2007;109:8–14. - PubMed
-
- McMichael G, Bainbridge MN, Haan E, et al. Whole‐exome sequencing points to considerable genetic heterogeneity of cerebral palsy. Mol Psych 2015;20:176–182. - PubMed
-
- MacLennan AH, Thompson SC, Gecz J. Cerebral palsy: causes, pathways, and the role of genetic variants. Am J Obstet Gynecol 2015;213:779–788. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

