EuPathDB: The Eukaryotic Pathogen Genomics Database Resource
- PMID: 29761457
- PMCID: PMC7124890
- DOI: 10.1007/978-1-4939-7737-6_5
EuPathDB: The Eukaryotic Pathogen Genomics Database Resource
Abstract
Fighting infections and developing novel drugs and vaccines requires advanced knowledge of pathogen's biology. Readily accessible genomic, functional genomic, and population data aids biological and translational discovery. The Eukaryotic Pathogen Database Resources ( http://eupathdb.org ) are data mining resources that support hypothesis driven research by facilitating the discovery of meaningful biological relationships from large volumes of data. The resource encompasses 13 sites that support over 170 species including pathogenic protists, oomycetes, and fungi as well as evolutionarily related nonpathogenic species. EuPathDB integrates preanalyzed data with advanced search capabilities, data visualization, analysis tools and a comprehensive record system in a graphical interface that does not require prior computational skills. This chapter describes guiding concepts common across EuPathDB sites and illustrates the powerful data mining capabilities of some of the available tools and features.
Keywords: Bioinformatics; Fungi; Genomics; Orthology; Parasite; Pathogen; Proteomics; Sequence analysis; Transcriptomics.
Figures





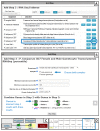
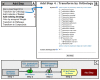
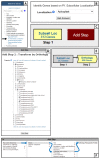

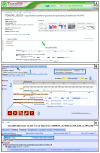



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

