Using FlyBase to Find Functionally Related Drosophila Genes
- PMID: 29761468
- PMCID: PMC5996772
- DOI: 10.1007/978-1-4939-7737-6_16
Using FlyBase to Find Functionally Related Drosophila Genes
Abstract
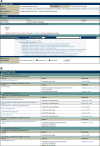
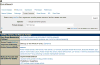
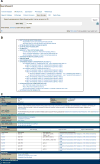
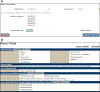
For more than 25 years, FlyBase ( flybase.org ) has served as an online database of biological information on the genus Drosophila, concentrating on the model organism D. melanogaster. Traditionally, FlyBase data have been organized and presented at a gene-by-gene level, which remains a useful perspective when the object of interest is a specific gene or gene product. However, in the modern era of a fully sequenced genome and an increasingly characterized proteome, it is often desirable to compile and analyze lists of genes related by a common function. This may be achieved in FlyBase by searching for genes annotated with relevant Gene Ontology (GO) terms and/or protein domain data. In addition, FlyBase provides preassembled lists of functionally related D. melanogaster genes within "Gene Group" reports. These are compiled manually from the published literature or expert databases and greatly facilitate access to, and analysis of, established gene sets. This chapter describes protocols to produce lists of functionally related genes in FlyBase using GO annotations, protein domain data and the Gene Groups resource, and provides guidance and advice for their further analysis and processing.
Keywords: D. melanogaster; Database; Drosophila; FlyBase; Functionally related genes; Gene Ontology; Gene group; Protein domain.
Figures


References
-
- Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, Falls K, Goodman JL, Hu Y, Ponting L, Schroeder AJ, Strelets VB, Thurmond J, Zhou P, FlyBase Consortium FlyBase at 25: looking to the future. Nucleic Acids Res. 2017;45(D1):D663–D671. https://doi.org/10.1093/nar/gkw1016. - DOI - PMC - PubMed
-
- Marygold SJ, Crosby MA, Goodman JL, FlyBase Consortium Using FlyBase, a database of Drosophila genes and genomes. Methods Mol Biol. 2016;1478:1–31. https://doi.org/10.1007/978-1-4939-6371-3_1. - DOI - PMC - PubMed
-
- The Gene Ontology C. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45(D1):D331–D338. https://doi.org/10.1093/nar/gkw1108. - DOI - PMC - PubMed
-
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztanyi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SC, Wu CH, Xenarios I, Yeh LS, Young SY, Mitchell AL. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45(D1):D190–D199. https://doi.org/10.1093/nar/gkw1107. - DOI - PMC - PubMed
-
- Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12(5):449–462. https://doi.org/10.1093/bib/bbr042. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

