Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins
- PMID: 29761785
- PMCID: PMC5953540
- DOI: 10.7554/eLife.33953
Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins
Abstract
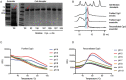
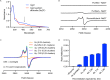
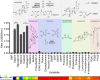
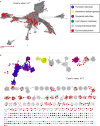
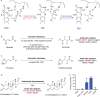
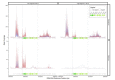
Although the human gut microbiome plays a prominent role in xenobiotic transformation, most of the genes and enzymes responsible for this metabolism are unknown. Recently, we linked the two-gene 'cardiac glycoside reductase' (cgr) operon encoded by the gut Actinobacterium Eggerthella lenta to inactivation of the cardiac medication and plant natural product digoxin. Here, we compared the genomes of 25 E. lenta strains and close relatives, revealing an expanded 8-gene cgr-associated gene cluster present in all digoxin metabolizers and absent in non-metabolizers. Using heterologous expression and in vitro biochemical characterization, we discovered that a single flavin- and [4Fe-4S] cluster-dependent reductase, Cgr2, is sufficient for digoxin inactivation. Unexpectedly, Cgr2 displayed strict specificity for digoxin and other cardenolides. Quantification of cgr2 in gut microbiomes revealed that this gene is widespread and conserved in the human population. Together, these results demonstrate that human-associated gut bacteria maintain specialized enzymes that protect against ingested plant toxins.
Trial registration: ClinicalTrials.gov NCT03022682 NCT01967563 NCT01105143.
Keywords: Eggerthella lenta; digoxin; enzyme; gut microbiome; infectious disease; microbiology; xenobiotic.
© 2018, Koppel et al.
Conflict of interest statement
NK, JB, MP No competing interests declared, PT PJT is on the scientific advisory board for Seres Therapeutics, WholeBiome, and Kaleido, and has active research funding from Medimmune. In the past year PJT has consulted for GLG and MEDAcorp. EB EPB is a consultant for Merck Research Labs, Novartis, and Kintai Therapeutics.
Figures














References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

