Lipidome Evolution in Mammalian Tissues
- PMID: 29762743
- PMCID: PMC6063302
- DOI: 10.1093/molbev/msy097
Lipidome Evolution in Mammalian Tissues
Abstract
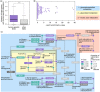
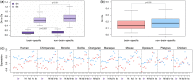
Lipids are essential structural and functional components of cells. Little is known, however, about the evolution of lipid composition in different tissues. Here, we report a large-scale analysis of the lipidome evolution in six tissues of 32 species representing primates, rodents, and bats. While changes in genes' sequence and expression accumulate proportionally to the phylogenetic distances, <2% of the lipidome evolves this way. Yet, lipids constituting this 2% cluster in specific functions shared among all tissues. Among species, human show the largest amount of species-specific lipidome differences. Many of the uniquely human lipidome features localize in the brain cortex and cluster in specific pathways implicated in cognitive disorders.
Figures




Similar articles
-
The LUX Score: A Metric for Lipidome Homology.PLoS Comput Biol. 2015 Sep 22;11(9):e1004511. doi: 10.1371/journal.pcbi.1004511. eCollection 2015. PLoS Comput Biol. 2015. PMID: 26393792 Free PMC article.
-
Lipidome determinants of maximal lifespan in mammals.Sci Rep. 2017 Jan 31;7(1):5. doi: 10.1038/s41598-017-00037-7. Sci Rep. 2017. PMID: 28127055 Free PMC article.
-
The Effects of Long-Term Saturated Fat Enriched Diets on the Brain Lipidome.PLoS One. 2016 Dec 1;11(12):e0166964. doi: 10.1371/journal.pone.0166964. eCollection 2016. PLoS One. 2016. PMID: 27907021 Free PMC article.
-
Use of long sequence alignments to study the evolution and regulation of mammalian globin gene clusters.Mol Biol Evol. 1993 Jan;10(1):73-102. doi: 10.1093/oxfordjournals.molbev.a039991. Mol Biol Evol. 1993. PMID: 8383794 Review.
-
Not all brains are made the same: new views on brain scaling in evolution.Brain Behav Evol. 2011;78(1):22-36. doi: 10.1159/000327318. Epub 2011 Jun 17. Brain Behav Evol. 2011. PMID: 21691045 Review.
Cited by
-
Differences in lipidome and metabolome organization of prefrontal cortex among human populations.Sci Rep. 2019 Dec 4;9(1):18348. doi: 10.1038/s41598-019-53762-6. Sci Rep. 2019. PMID: 31797944 Free PMC article.
-
Retroelement-Linked H3K4me1 Histone Tags Uncover Regulatory Evolution Trends of Gene Enhancers and Feature Quickly Evolving Molecular Processes in Human Physiology.Cells. 2019 Oct 8;8(10):1219. doi: 10.3390/cells8101219. Cells. 2019. PMID: 31597351 Free PMC article.
-
A Comprehensive Profiling of Cellular Sphingolipids in Mammalian Endothelial and Microglial Cells Cultured in Normal and High-Glucose Conditions.Cells. 2022 Sep 30;11(19):3082. doi: 10.3390/cells11193082. Cells. 2022. PMID: 36231042 Free PMC article.
-
Aging, VO2 max, entropy, and COVID-19.Indian J Pharmacol. 2022 Jan-Feb;54(1):58-62. doi: 10.4103/ijp.ijp_442_21. Indian J Pharmacol. 2022. PMID: 35343209 Free PMC article. Review.
-
Age-Related Changes in Lipidome of Rat Frontal Cortex and Cerebellum Are Partially Reversed by Methionine Restriction Applied in Old Age.Int J Mol Sci. 2021 Nov 20;22(22):12517. doi: 10.3390/ijms222212517. Int J Mol Sci. 2021. PMID: 34830402 Free PMC article.
References
-
- Bozek K, Wei Y, Yan Z, Liu X, Xiong J, Sugimoto M, Tomita M, Pääbo S, Sherwood CC, Hof Patrick R, et al. . 2015. Organization and evolution of brain lipidome revealed by large-scale analysis of human, chimpanzee, macaque, and mouse tissues. Neuron 854:695–702. - PubMed
-
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. . 2011. The evolution of gene expression levels in mammalian organs. Nature 4787369:343–348. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

