Molecular, Pathological, Radiological, and Immune Profiling of Non-brainstem Pediatric High-Grade Glioma from the HERBY Phase II Randomized Trial
- PMID: 29763623
- PMCID: PMC5956280
- DOI: 10.1016/j.ccell.2018.04.004
Molecular, Pathological, Radiological, and Immune Profiling of Non-brainstem Pediatric High-Grade Glioma from the HERBY Phase II Randomized Trial
Abstract
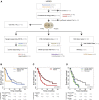
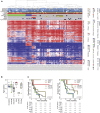
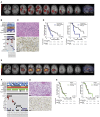
The HERBY trial was a phase II open-label, randomized, multicenter trial evaluating bevacizumab (BEV) in addition to temozolomide/radiotherapy in patients with newly diagnosed non-brainstem high-grade glioma (HGG) between the ages of 3 and 18 years. We carried out comprehensive molecular analysis integrated with pathology, radiology, and immune profiling. In post-hoc subgroup analysis, hypermutator tumors (mismatch repair deficiency and somatic POLE/POLD1 mutations) and those biologically resembling pleomorphic xanthoastrocytoma ([PXA]-like, driven by BRAF_V600E or NF1 mutation) had significantly more CD8+ tumor-infiltrating lymphocytes, and longer survival with the addition of BEV. Histone H3 subgroups (hemispheric G34R/V and midline K27M) had a worse outcome and were immune cold. Future clinical trials will need to take into account the diversity represented by the term "HGG" in the pediatric population.
Trial registration: ClinicalTrials.gov NCT01390948.
Keywords: CD8; H3F3A; MAPK; hypermutator; immune; pediatric high-grade glioma.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Bautista F., Paci A., Minard-Colin V., Dufour C., Grill J., Lacroix L., Varlet P., Valteau-Couanet D., Geoerger B. Vemurafenib in pediatric patients with BRAFV600E mutated high-grade gliomas. Pediatr. Blood Cancer. 2014;61:1101–1103. - PubMed
-
- Benesch M., Windelberg M., Sauseng W., Witt V., Fleischhack G., Lackner H., Gadner H., Bode U., Urban C. Compassionate use of bevacizumab (Avastin) in children and young adults with refractory or recurrent solid tumors. Ann. Oncol. 2008;19:807–813. - PubMed
-
- Bouffet E., Larouche V., Campbell B.B., Merico D., de Borja R., Aronson M., Durno C., Krueger J., Cabric V., Ramaswamy V. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol. 2016;34:2206–2211. - PubMed
-
- Brouwer-Visser J., Cheng W.Y., Bauer-Mehren A., Maisel D., Lechner K., Andersson E., Dudley J.T., Milletti F. Regulatory T-cell genes drive altered immune microenvironment in adult solid cancers and allow for immune contextual patient subtyping. Cancer Epidemiol. Biomarkers Prev. 2018;27:103–112. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

