The GSK3 Signaling Axis Regulates Adaptive Glutamine Metabolism in Lung Squamous Cell Carcinoma
- PMID: 29763624
- PMCID: PMC6451645
- DOI: 10.1016/j.ccell.2018.04.002
The GSK3 Signaling Axis Regulates Adaptive Glutamine Metabolism in Lung Squamous Cell Carcinoma
Abstract
Altered metabolism is a hallmark of cancer growth, forming the conceptual basis for development of metabolic therapies as cancer treatments. We performed in vivo metabolic profiling and molecular analysis of lung squamous cell carcinoma (SCC) to identify metabolic nodes for therapeutic targeting. Lung SCCs adapt to chronic mTOR inhibition and suppression of glycolysis through the GSK3α/β signaling pathway, which upregulates glutaminolysis. Phospho-GSK3α/β protein levels are predictive of response to single-therapy mTOR inhibition while combinatorial treatment with the glutaminase inhibitor CB-839 effectively overcomes therapy resistance. In addition, we identified a conserved metabolic signature in a broad spectrum of hypermetabolic human tumors that may be predictive of patient outcome and response to combined metabolic therapies targeting mTOR and glutaminase.
Keywords: CB-839; GLS; GSK3α/β; PET imaging; glutaminolysis; glycolysis; lung squamous cell carcinoma; mTOR; metabolic signature.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures
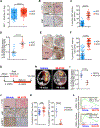

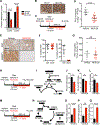

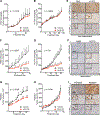


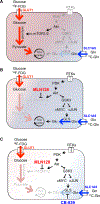
Comment in
-
Targeting Therapy Resistance: When Glutamine Catabolism Becomes Essential.Cancer Cell. 2018 May 14;33(5):795-797. doi: 10.1016/j.ccell.2018.04.009. Cancer Cell. 2018. PMID: 29763620 Free PMC article.
References
-
- Berghmans T, Dusart M, Paesmans M, Hossein-Foucher C, Buvat I, Castaigne C, Scherpereel A, Mascaux C, Moreau M, Roelandts M, et al. (2008). Primary tumor standardized uptake value (SUVmax) measured on fluorodeoxyglucose positron emission tomography (FDG-PET) is of prognostic value for survival in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis (MA) by the European Lung Cancer Working Party for the IASLC Lung Cancer Staging Project. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 3, 6–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

