Curative Ex Vivo Hepatocyte-Directed Gene Editing in a Mouse Model of Hereditary Tyrosinemia Type 1
- PMID: 29764210
- PMCID: PMC6247987
- DOI: 10.1089/hum.2017.252
Curative Ex Vivo Hepatocyte-Directed Gene Editing in a Mouse Model of Hereditary Tyrosinemia Type 1
Abstract
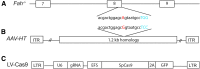
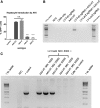
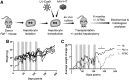
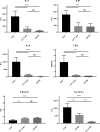
Hereditary tyrosinemia type 1 (HT1) is an autosomal recessive disorder caused by deficiency of fumarylacetoacetate hydrolase (FAH). It has been previously shown that ex vivo hepatocyte-directed gene therapy using an integrating lentiviral vector to replace the defective Fah gene can cure liver disease in small- and large-animal models of HT1. This study hypothesized that ex vivo hepatocyte-directed gene editing using CRISPR/Cas9 could be used to correct a mouse model of HT1, in which a single point mutation results in loss of FAH function. To achieve high transduction efficiencies of primary hepatocytes, this study utilized a lentiviral vector (LV) to deliver both the Streptococcus pyogenes Cas9 nuclease and target guide RNA (LV-Cas9) and an adeno-associated virus (AAV) vector to deliver a 1.2 kb homology template (AAV-HT). Cells were isolated from Fah-/- mice and cultured in the presence of LV and AAV vectors. Transduction of cells with LV-Cas9 induced significant indels at the target locus, and correction of the point mutation in Fah-/- cells ex vivo using AAV-HT was completely dependent on LV-Cas9. Next, hepatocytes transduced ex vivo by LV-Cas9 and AAV-HT were transplanted into syngeneic Fah-/- mice that had undergone a two-thirds partial hepatectomy or sham hepatectomy. Mice were cycled on/off the protective drug 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC) to stimulate expansion of corrected cells. All transplanted mice became weight stable off NTBC. However, a significant improvement was observed in weight stability off NTBC in animals that received partial hepatectomy. After 6 months, mice were euthanized, and thorough biochemical and histological examinations were performed. Biochemical markers of liver injury were significantly improved over non-transplanted controls. Histological examination of mice revealed normal tissue architecture, while immunohistochemistry showed robust repopulation of recipient animals with FAH+ cells. In summary, this is the first report of ex vivo hepatocyte-directed gene repair using CRISPR/Cas9 to demonstrate curative therapy in an animal model of liver disease.
Keywords: CRISPR/Cas9; gene therapy; hepatocytes; hereditary tyrosinemia type 1; metabolic liver disease.
Conflict of interest statement
The authors declare no competing financial interests exist.
Figures

Similar articles
-
Curative ex vivo liver-directed gene therapy in a pig model of hereditary tyrosinemia type 1.Sci Transl Med. 2016 Jul 27;8(349):349ra99. doi: 10.1126/scitranslmed.aaf3838. Sci Transl Med. 2016. PMID: 27464750 Free PMC article.
-
Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats.J Biol Chem. 2018 May 4;293(18):6883-6892. doi: 10.1074/jbc.RA117.000347. Epub 2018 Mar 5. J Biol Chem. 2018. PMID: 29507093 Free PMC article.
-
Autologous Gene and Cell Therapy Provides Safe and Long-Term Curative Therapy in A Large Pig Model of Hereditary Tyrosinemia Type 1.Cell Transplant. 2019 Jan;28(1):79-88. doi: 10.1177/0963689718814188. Epub 2018 Nov 26. Cell Transplant. 2019. PMID: 30477316 Free PMC article.
-
Nuclease-Mediated Gene Therapies for Inherited Metabolic Diseases of the Liver.Yale J Biol Med. 2017 Dec 19;90(4):553-566. eCollection 2017 Dec. Yale J Biol Med. 2017. PMID: 29259521 Free PMC article. Review.
-
Biochemical and Clinical Aspects of Hereditary Tyrosinemia Type 1.Adv Exp Med Biol. 2017;959:9-21. doi: 10.1007/978-3-319-55780-9_2. Adv Exp Med Biol. 2017. PMID: 28755181 Review.
Cited by
-
Electroporation-Mediated Delivery of Cas9 Ribonucleoproteins Results in High Levels of Gene Editing in Primary Hepatocytes.CRISPR J. 2022 Jun;5(3):397-409. doi: 10.1089/crispr.2021.0134. Epub 2022 Mar 2. CRISPR J. 2022. PMID: 35238624 Free PMC article.
-
In vitro Studies of Transendothelial Migration for Biological and Drug Discovery.Front Med Technol. 2020 Nov 16;2:600616. doi: 10.3389/fmedt.2020.600616. eCollection 2020. Front Med Technol. 2020. PMID: 35047883 Free PMC article. Review.
-
Novel Gene-Correction-Based Therapeutic Modalities for Monogenic Liver Disorders.Bioengineering (Basel). 2022 Aug 15;9(8):392. doi: 10.3390/bioengineering9080392. Bioengineering (Basel). 2022. PMID: 36004917 Free PMC article. Review.
-
Strategies for the CRISPR-Based Therapeutics.Trends Pharmacol Sci. 2020 Jan;41(1):55-65. doi: 10.1016/j.tips.2019.11.006. Epub 2019 Dec 17. Trends Pharmacol Sci. 2020. PMID: 31862124 Free PMC article. Review.
-
Mutational spectrum of Mexican patients with tyrosinemia type 1: In silico modeling and predicted pathogenic effect of a novel missense FAH variant.Mol Genet Genomic Med. 2019 Dec;7(12):e937. doi: 10.1002/mgg3.937. Epub 2019 Sep 30. Mol Genet Genomic Med. 2019. PMID: 31568711 Free PMC article.
References
-
- Grompe M. The pathophysiology and treatment of hereditary tyrosinemia type 1. Semin Liver Dis 2001;21:563–571 - PubMed
-
- Grompe M, Lindstedt S, al-Dhalimy M, et al. . Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat Genet 1995;10:453–460 - PubMed
-
- Sniderman King L, Trahms C, Scott CR. Tyrosinemia type I. In: Adam MP, Ardinger HH, Pagon RA, et al., eds. GeneReviews®. Seattle, WA: University of Washington, 1993
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

