Phylogenetic signal from rearrangements in 18 Anopheles species by joint scaffolding extant and ancestral genomes
- PMID: 29764366
- PMCID: PMC5954271
- DOI: 10.1186/s12864-018-4466-7
Phylogenetic signal from rearrangements in 18 Anopheles species by joint scaffolding extant and ancestral genomes
Abstract
Background: Genomes rearrangements carry valuable information for phylogenetic inference or the elucidation of molecular mechanisms of adaptation. However, the detection of genome rearrangements is often hampered by current deficiencies in data and methods: Genomes obtained from short sequence reads have generally very fragmented assemblies, and comparing multiple gene orders generally leads to computationally intractable algorithmic questions.
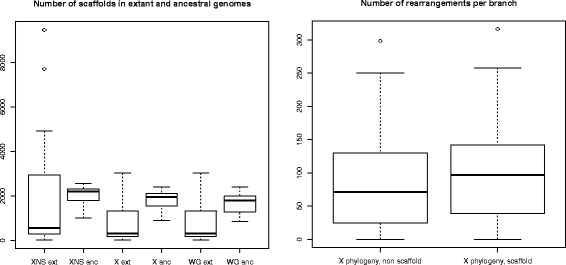
Results: We present a computational method, ADSEQ, which, by combining ancestral gene order reconstruction, comparative scaffolding and de novo scaffolding methods, overcomes these two caveats. ADSEQ provides simultaneously improved assemblies and ancestral genomes, with statistical supports on all local features. Compared to previous comparative methods, it runs in polynomial time, it samples solutions in a probabilistic space, and it can handle a significantly larger gene complement from the considered extant genomes, with complex histories including gene duplications and losses. We use ADSEQ to provide improved assemblies and a genome history made of duplications, losses, gene translocations, rearrangements, of 18 complete Anopheles genomes, including several important malaria vectors. We also provide additional support for a differentiated mode of evolution of the sex chromosome and of the autosomes in these mosquito genomes.
Conclusions: We demonstrate the method's ability to improve extant assemblies accurately through a procedure simulating realistic assembly fragmentation. We study a debated issue regarding the phylogeny of the Gambiae complex group of Anopheles genomes in the light of the evolution of chromosomal rearrangements, suggesting that the phylogenetic signal they carry can differ from the phylogenetic signal carried by gene sequences, more prone to introgression.
Keywords: Comparative genomics; Mosquito genomics; Scaffolding.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

