High serum levels of pregenomic RNA reflect frequently failing reverse transcription in hepatitis B virus particles
- PMID: 29764511
- PMCID: PMC5952638
- DOI: 10.1186/s12985-018-0994-7
High serum levels of pregenomic RNA reflect frequently failing reverse transcription in hepatitis B virus particles
Abstract
Background: Hepatocytes infected by hepatitis B virus (HBV) produce different HBV RNA species, including pregenomic RNA (pgRNA), which is reverse transcribed during replication. Particles containing HBV RNA are present in serum of infected individuals, and quantification of this HBV RNA could be clinically useful.
Methods: In a retrospective study of 95 patients with chronic HBV infection, we characterised HBV RNA in serum in terms of concentration, particle association and sequence. HBV RNA was detected by real-time PCR at levels almost as high as HBV DNA.
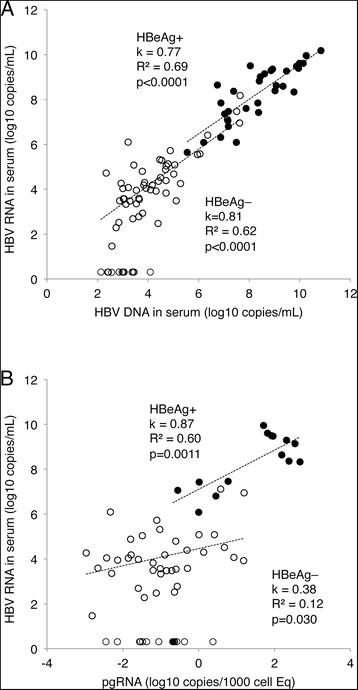
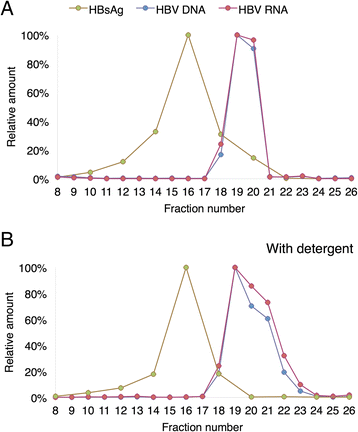
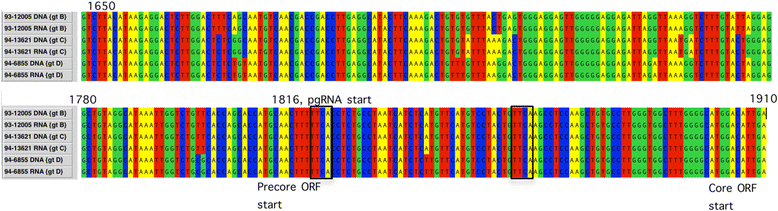
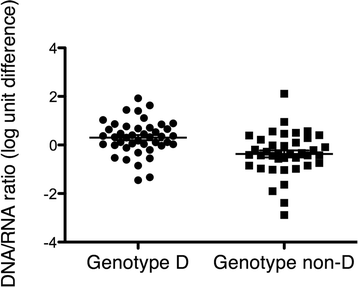
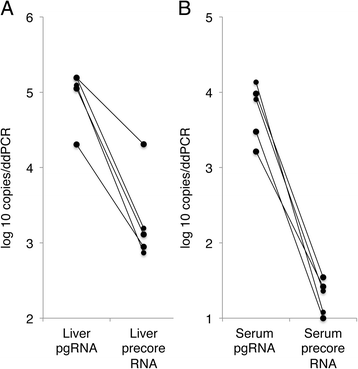
Results: The HBV RNA was protected from RNase and it was found in particles of similar density as particles containing HBV DNA after fractionation on a Nycodenz gradient. Sequencing the epsilon region of the RNA did not reveal mutations that would preclude its binding to the viral polymerase before encapsidation. Specific quantification of precore RNA and pgRNA by digital PCR showed almost seven times lower ratio of precore RNA/pgRNA in serum than in liver tissue, which corresponds to poorer encapsidation of this RNA as compared with pgRNA. The serum ratio between HBV DNA and HBV RNA was higher in genotype D as compared with other genotypes.
Conclusions: The results suggest that HBV RNA in serum is present in viral particles with failing reverse transcription activity, which are produced at almost as high rates as viral particles containing DNA. The results encourage further studies of the mechanisms by which these particles are produced, the impact of genotype, and the potential clinical utility of quantifying HBV RNA in serum.
Conflict of interest statement
Ethics approval and consent to participate
The studies were conducted in accordance with the Declaration of Helsinki and approved by the Regional Ethical Review Board in Gothenburg, Sweden. An informed consent was obtained from each patient.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Intracellular Trafficking of HBV Particles.Cells. 2020 Sep 2;9(9):2023. doi: 10.3390/cells9092023. Cells. 2020. PMID: 32887393 Free PMC article. Review.
-
Relative Abundance of Integrant-Derived Viral RNAs in Infected Tissues Harvested from Chronic Hepatitis B Virus Carriers.J Virol. 2018 Apr 27;92(10):e02221-17. doi: 10.1128/JVI.02221-17. Print 2018 May 15. J Virol. 2018. PMID: 29491161 Free PMC article.
-
Mechanisms downstream of reverse transcription reduce serum levels of HBV DNA but not of HBsAg in chronic hepatitis B virus infection.Virol J. 2015 Dec 9;12:213. doi: 10.1186/s12985-015-0447-5. Virol J. 2015. PMID: 26645241 Free PMC article.
-
Serum HBV DNA plus RNA shows superiority in reflecting the activity of intrahepatic cccDNA in treatment-naïve HBV-infected individuals.J Clin Virol. 2018 Feb-Mar;99-100:71-78. doi: 10.1016/j.jcv.2017.12.016. Epub 2018 Jan 6. J Clin Virol. 2018. PMID: 29353073
-
Structure and function of the encapsidation signal of hepadnaviridae.J Viral Hepat. 1998 Nov;5(6):357-67. doi: 10.1046/j.1365-2893.1998.00124.x. J Viral Hepat. 1998. PMID: 9857345 Review.
Cited by
-
Current tests for diagnosis of hepatitis B virus infection and immune responses of HBV-related HCC.Front Oncol. 2023 Nov 28;13:1185142. doi: 10.3389/fonc.2023.1185142. eCollection 2023. Front Oncol. 2023. PMID: 38090482 Free PMC article. Review.
-
Clinical Implications of Serum Hepatitis B Virus Pregenomic RNA Kinetics in Chronic Hepatitis B Patients Receiving Antiviral Treatment and Those Achieving HBsAg Loss.Microorganisms. 2021 May 26;9(6):1146. doi: 10.3390/microorganisms9061146. Microorganisms. 2021. PMID: 34073483 Free PMC article.
-
Dynamics of Serum Pregenome RNA in Chronic Hepatitis B Patients Receiving 96-Month Nucleos(t)ide Analog Therapy.Front Med (Lausanne). 2022 Feb 28;9:787770. doi: 10.3389/fmed.2022.787770. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35295596 Free PMC article.
-
Utility of novel viral and immune markers in predicting HBV treatment endpoints: A systematic review of treatment discontinuation studies.JHEP Rep. 2023 Mar 8;5(6):100720. doi: 10.1016/j.jhepr.2023.100720. eCollection 2023 Jun. JHEP Rep. 2023. PMID: 37138673 Free PMC article.
-
Serum Hepatitis B Virus RNA: A New Potential Biomarker for Chronic Hepatitis B Virus Infection.Hepatology. 2019 Apr;69(4):1816-1827. doi: 10.1002/hep.30325. Epub 2019 Mar 20. Hepatology. 2019. PMID: 30362148 Free PMC article. Review.
References
-
- Thompson AJV, Nguyen T, Iser D, Ayres A, Jackson K, Littlejohn M, et al. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology. 2010;51:1933–1944. doi: 10.1002/hep.23571. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

