HP1γ Promotes Lung Adenocarcinoma by Downregulating the Transcription-Repressive Regulators NCOR2 and ZBTB7A
- PMID: 29764865
- PMCID: PMC6443096
- DOI: 10.1158/0008-5472.CAN-17-3571
HP1γ Promotes Lung Adenocarcinoma by Downregulating the Transcription-Repressive Regulators NCOR2 and ZBTB7A
Abstract
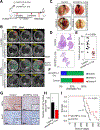
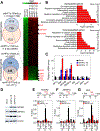
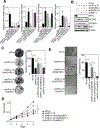
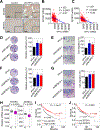
Lung adenocarcinoma is a major form of lung cancer, which is the leading cause of cancer death. Histone methylation reader proteins mediate the effect of histone methylation, a hallmark of epigenetic and transcriptional regulation of gene expression. However, their roles in lung adenocarcinoma are poorly understood. Here, our bioinformatic screening and analysis in search of a lung adenocarcinoma-promoting histone methylation reader protein show that heterochromatin protein 1γ (HP1γ; also called CBX3) is among the most frequently overexpressed and amplified histone reader proteins in human lung adenocarcinoma, and that high HP1γ mRNA levels are associated with poor prognosis in patients with lung adenocarcinoma. In vivo depletion of HP1γ reduced K-RasG12D-driven lung adenocarcinoma and lengthened survival of mice bearing K-RasG12D-induced lung adenocarcinoma. HP1γ and its binding activity to methylated histone H3 lysine 9 were required for the proliferation, colony formation, and migration of lung adenocarcinoma cells. HP1γ directly repressed expression of the transcription-repressive regulators NCOR2 and ZBTB7A. Knockdown of NCOR2 or ZBTB7A significantly restored defects in proliferation, colony formation, and migration in HP1γ-depleted lung adenocarcinoma cells. Low NCOR2 or ZBTB7A mRNA levels were associated with poor prognosis in patients with lung adenocarcinoma and correlated with high HP1γ mRNA levels in lung adenocarcinoma samples. NCOR2 and ZBTB7A downregulated expression of tumor-promoting factors such as ELK1 and AXL, respectively. These findings highlight the importance of HP1γ and its reader activity in lung adenocarcinoma tumorigenesis and reveal a unique lung adenocarcinoma-promoting mechanism in which HP1γ downregulates NCOR2 and ZBTB7A to enhance expression of protumorigenic genes.Significance: Direct epigenetic repression of the transcription-repressive regulators NCOR2 and ZBTB7A by the histone reader protein HP1γ leads to activation of protumorigenic genes in lung adenocarcinoma. Cancer Res; 78(14); 3834-48. ©2018 AACR.
©2018 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures



References
-
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

