SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis
- PMID: 29764982
- PMCID: PMC5983216
- DOI: 10.15252/embj.201797499
SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis
Abstract
Proper floral patterning, including the number and position of floral organs in most plant species, is tightly controlled by the precise regulation of the persistence and size of floral meristems (FMs). In Arabidopsis, two known feedback pathways, one composed of WUSCHEL (WUS) and CLAVATA3 (CLV3) and the other composed of AGAMOUS (AG) and WUS, spatially and temporally control floral stem cells, respectively. However, mounting evidence suggests that other factors, including phytohormones, are also involved in floral meristem regulation. Here, we show that the boundary gene SUPERMAN (SUP) bridges floral organogenesis and floral meristem determinacy in another pathway that involves auxin signaling. SUP interacts with components of polycomb repressive complex 2 (PRC2) and fine-tunes local auxin signaling by negatively regulating the expression of the auxin biosynthesis genes YUCCA1/4 (YUC1/4). In sup mutants, derepressed local YUC1/4 activity elevates auxin levels at the boundary between whorls 3 and 4, which leads to an increase in the number and the prolonged maintenance of floral stem cells, and consequently an increase in the number of reproductive organs. Our work presents a new floral meristem regulatory mechanism, in which SUP, a boundary gene, coordinates floral organogenesis and floral meristem size through fine-tuning auxin biosynthesis.
Keywords: SUPERMAN; H3K27me3; auxin; floral meristem; floral organogenesis; polycomb repressive complexes.
© 2018 The Authors.
Figures
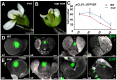
- A, B
wus‐1 (A) and sup‐5 wus‐1 (B) mutant flowers with one stamen and without carpels. Scale bars, 1 mm.
- C
The comparison of the number of cells with the stem cell marker pCLV3::GFP‐ER signals in WT and sup‐5. The numbers of cells with the signals were counted based on the z‐stack images. From stage 4 (s4) onwards, the sup‐5 floral buds showed increased numbers of CLV3‐expressing stem cells compared with those of WT. Error bars indicate s.d. of 12–15 samples; two‐tailed Student's t‐test, *P < 0.05.
- D, E
The pCLV3::GFP‐ER (green) in WT (D) and sup‐5 (E) floral buds at different floral stages. Scale bars, 20 μm.

- A
Expression of WUS and CLV3 (in floral buds of approximately stage 6) 5 days after treatment of ap1 cal p35S::AP1‐GR sup‐5 inflorescences with 1 μM DEX. The relative values to equally treated ap1 cal p35S::AP1‐GR plants are shown. Error bars indicate standard errors from qRT–PCR experiments of four biological repeats. P = 0.028 and 0.031 for WUS and CLV3 based on a Student's t‐test, respectively.
- B, C
The pCLV3::GFP‐ER (green) can be detected in a sup‐5 floral bud (C) but not in WT (B) at a floral stage later than 6. Scale bars, 20 μm.

- A, B
The expression of pSTM::CFP‐N7 (light blue) in WT and sup‐5 inflorescence. In both WT and sup‐5, the STM‐VENUS fusion protein was highly expressed in the center of floral buds younger than stage 5 as well as in the boundary regions of sepals.
- C, D
A stem cell marker pSTM::STM‐VENUS (red) in WT (C) and sup‐5 (D) floral buds at the stages 5 and 6. From late stage 5 (s5), STM was reduced in the regions with developing stamens. The STM expression domain in FM centers is relatively larger in sup‐5 (1,680 ± 167 μm2, n = 15) than that in WT (1,150 ± 160 μm2, n = 16). P < 0.05 based on a Student's t‐test. Dashed lines mark the FM regions and whorl 3/4 boundary regions with STM‐VENUS.
- E, F
The expression of pSTM::CFP‐N7 (light blue) only remains expressed at the carpel margins/placenta region in the WT at stage 7 (s7) and greatly diminished at stage 9 (s9) (E). Conversely, in sup‐5, STM is expressed in a larger domain, which encompasses the FM that keeps proliferating as well as in emerging extra stamen primordia up to stage later than 10 (s10+) (F).

pCUC2::3xVENUS‐N7 in WT floral buds at stages 3‐6. CUC2 was highly expressed in the floral buds of stage 3, and its expression started to be constrained to the boundary regions between the sepals after stage 3. At stage 5 and stage 6 floral buds, CUC2 was only detected in the small boundary regions at the bottom of the sepals.
pCUC2::3xVENUS‐N7 in sup‐5 floral buds at stages 3–6. At stage 3, the CUC2 expression pattern was similar to that in WT. From stage 4, the CUC2 expression pattern in sup‐5 started to show differences from that of WT. CUC2 expression was detected in FM region of at stage 4. At stages 5 and 6, the CUC2 expression was still relatively high but was more concentrated at the bottom of the developing sepals as well as in the broad FM regions. Dashed lines mark the regions with CUC2‐3xVENUS‐N7 in the flower center. “b” indicates the boundary region between sepals; “fm” indicates the floral meristem region in the center.
A cross section of a stage 4 WT floral bud showing a ring of CUC2‐3xVENUS‐N7 signal at the whorl 3/4 boundary.
A cross section of a stage 4 sup‐5 floral bud showing the additional signal of CUC2‐3xVENUS‐N7 in the FM center.

- A, B
Activity of the auxin marker pDR5rev::2xGFP‐N7 in stage 4 (s4) floral buds of WT (A) and sup‐1 (B). In WT flowers, the fluorescence signal was detected mostly at the tips of the sepals and at the sites (p, marked with the arrows) where the petal primordia would emerge at later stages (A). In sup‐1 flowers, strong reporter activity was additionally detected at the whorl 3/4 boundary.
- C, D
Activity of the auxin reporter DII‐VENUS (green) in stage 4 floral buds of WT (C) and sup‐1 (D). In WT flowers, fluorescence signals were detected at the whorl 3/4 boundary but were absent at the center of the FM and the tips of the sepals (C). In contrast, DII‐VENUS signals were absent at the whorl 3/4 boundary but were detected in the FM region (D).
- E, F
sup‐5 flowers after treatment with the anti‐auxin PCIP (F) and mock solutions (E). While the mock treatment did not affect sup‐5 flowers (E), treatment with PCIB strongly rescued both carpels and stamen numbers.
- G
The statistical analysis indicated that PCIB treatment strongly rescued stage 4–5 floral buds, which took approximately 9–10 days to anthesis; the floral buds of stage 6 were best rescued in terms of carpel morphology. Error bars indicate s.d. of 20 flowers from around 10 individual plants; two‐tailed Student's t‐test, *P < 0.05.
- H–J
pSUP::iaaH transgenic flowers with the IAM treatment mimicked the various sup‐like phenotypes, including increased stamen numbers and defective carpels, as shown in the SEM images of the flowers (H, I). Wild‐type plants (lacking pSUP:iaaH) were treated with IAM as a negative control. The number of free stamen, fusion stamen, and carpels was determined for a total of 20 flowers from 20 individual plants and is summarized in (J). Error bars indicate s.d.
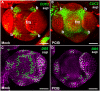
- A, B
The expression of pCUC2::CUC2‐3xVENUS‐N7 was induced in stage 4 floral buds of sup‐5 plants 4–6 h after treatment with PCIB (B) but after a mock treatment (A).
- C, D
The expression of pDR5rev::3xVENUS‐N7 was reduced in stage 4 floral buds of sup‐5 plants 4–6 h after treatment with PCIB (D) but not after a mock treatment (C).

REVIGO analysis of pathways significantly enriched among the down‐regulated genes. Each rectangle is a single cluster representative for the non‐redundant GO term, which are joined into “superclusters” of related terms, visualized with different colors. Size of the rectangles reflects the P‐value.
YUC1/4 and PIN3/4 are reduced in p35S::SUP‐GR inflorescences 2 and 4 h after treatment with 10 μM DEX. Error bars indicate s.d. of three biological replicates; two‐tailed Student's t‐test, *P < 0.05.
Expression levels of YUC1/4 and PIN3/4 3 days after 1 μM DEX treatment in ap1 cal p35S::AP1‐GR sup‐5 and ap1 cal p35S::AP1‐GR. Expression of YUC1/4 was increased in the ap1 cal p35S::AP1‐GR sup‐5 background, while that of PIN3/4 was reduced. Error bars indicate s.d. of three biological replicates; two‐tailed Student's t‐test, *P < 0.05.
IAA levels in ap1 cal p35S::AP1‐GR sup‐5 and ap1 cal p35S::AP1‐GR 3 days after treatment with 1 μM DEX or a mock solution. The P‐value was calculated using one‐way ANOVA and standard errors from three biological replicates.
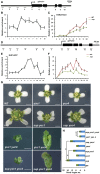
Schematic drawing of the YUC1 genome structure showing regions amplified by primer sets used for ChIP analyses.
ChIP binding assay of the SUP protein at the YUC1 genome. Relative enrichment of SUP‐GFP at the YUC1 locus, including the promoter and coding regions, was analyzed using the stage 4 floral buds from ap1 cal p35S::AP1‐GR sup‐5 pSUP::SUP‐GFP plants.
Relative enrichment of H3K27me3 at the YUC1 locus is decreased in the stage 4 floral buds of ap1 cal p35S::AP1‐GR sup‐5 plants relative to ap1 cal p35S::AP1‐GR.
Schematic drawing of the YUC4 gene structure showing regions amplified by primers used for ChIP analyses.
ChIP binding assay of the SUP protein at the YUC4 gene. Relative enrichment of SUP‐GFP at the YUC4 locus, including the promoter and coding regions, analyzed using the stage 4 floral buds of ap1 cal p35S::AP1‐GR sup‐5 pSUP::SUP‐GFP.
Relative enrichment of H3K27me3 is decreased at the YUC4 locus in stage 4 floral buds of ap1 cal p35S::AP1‐GR sup‐5 compared with that of the WT control. Error bars represent standard errors with three biological repeats.
yuc1 and yuc4 mutant alleles partially rescue the stamen and carpel number defects of sup‐5, while yuc1 yuc4 is epistatic to sup‐5. The yuc4 mutation results in reduced floral organ size while yuc4 appears WT‐like. Both yuc1 and yuc4 single mutant have no defects in floral organ numbers. The yuc1 yuc4 double mutant shows strong floral defects, namely a decreased numbers of stamen and carpel together with a reduction in floral organ size. The sup‐5 yuc1 yuc4 triple mutant shows a yuc1 yuc4‐like floral morphology. A typical flower is shown for WT, yuc1, yuc4, sup‐5, sup‐5 yuc1, sup‐5 yuc4, yuc1 yuc4, and sup‐5 yuc1 yuc4, respectively. For yuc1 yuc4 and sup‐5 yuc1 yuc4, the top view and side view are shown for the same flower. Scale bars: 1 mm.
The statistical analysis showed a reduction in total stamens and carpels in sup‐5 yuc1 (8.60 ± 1.33 for stamen, 2.13 ± 0.34 for carpel, n = 30), sup‐5 yuc4 (10.93 ± 1.74 for stamen, 2.4 ± 0.50 for carpel, n = 30) once compared with sup‐5 (14.00 ± 1.68 for stamen, 2.7 ± 0.75 for carpel, n = 30). *P < 0.05 based on Student's t‐test. There is no significant difference between yuc1 yuc4 (3.8 ± 0.77 for stamen, 1.25 ± 0.44 for carpel, n = 20) and sup‐5 yuc‐1 yuc4 (4.05 ± 0.70 for stamen, 1.05 ± 0.23 for carpel, n = 19) in floral organ number and floral organ size, P > 0.05 with a Student's t‐test.

- A, B
GUS staining of YUC1 full‐length reporter pYUC1::YUC1‐GUS in WT (A) and sup‐5 (B) inflorescences. The expression of pYUC1::YUC1‐GUS is weak at floral buds earlier than stage 5. At stage 5, GUS starts to be highly expressed at the sepals, especially at the bottom of sepals at the later stages. In sup‐5, pYUC1::YUC1‐GUS is expressed highly at the SUP expression domain as well as the FM region at the stage 4. Floral buds at stage 4 with GUS staining are shown as insets.
- C, D
GUS staining of YUC4 full‐length reporter pYUC4::YUC4‐GUS in WT (C) and sup‐5 (D) inflorescences. In WT, the expression of pYUC4::YUC4‐GUS is weak at the floral buds at earlier than stage 5. From stage 5, GUS is almost expressed in the whole floral buds (C). In sup‐5, pYUC4::YUC4‐GUS is highly expressed at the SUP expression domain at the floral buds of stage 4. Floral buds at stage 4 with GUS staining are shown as insets.
- E, F
GUS staining of YUC1 promoter reporter pYUC1::GUS was detected in almost all WT (E) and sup‐5 (F) young floral buds.
- G, H
GUS staining of YUC4 promoter reporter pYUC4::GUS was detected in almost all WT (G) and sup‐5 (H) young floral buds.

- A, B
The yeast two‐hybrid assay using a series of truncated SUP proteins with CLF and TFL2. Full‐length, three truncated SUP proteins and SUP proteins with the two versions of the abolished EAR motif (EARm1 and EARm2) were fused to a GAL4 DNA‐binding domain (BD). Schematic structures of the full‐length, truncated, or mutated SUP protein are shown in (B). The blue region indicates the zinc finger domain; the red regions indicate an intact EAR motif; and beige and black regions indicate mutated EAR motif, respectively. The truncated CLF without its C‐terminal SET domain (CLF) and the full‐length TFL2 was fused to the GAL4 activation domain (AD). Yeast colonies harboring these fusion constructs and/or empty vectors as indicated were grown on selective media of 2DO, 3DO, and 4DO. For CLF, yeast growth was only detected when the combination of the full‐length SUP and the truncated CLF was co‐transformed. None of the truncated SUP proteins or SUP proteins with the mutated EAF motif interacted with the truncated CLF. For TFL2, a short domain of SUP (amino acids 89–205) was sufficient for the interaction, and the mutation of EAR motifs did not affect the interaction with TFL2.
- C
Interaction between SUP and CLF as determined by co‐IP. Using stage 4 floral buds of ap1 cal p35S::AP1‐GR pSUP::SUP‐GFP pCLF::HA‐CLF or of ap1 cal p35S:: AP1‐GR pCLF::HA‐CLF plants, total proteins (input) were subjected to immunoprecipitation with anti‐GFP‐conjugated beads. Immunoblotting analysis with another anti‐GFP and anti‐HA antibody were performed to detect SUP‐GFP (to test pull‐down efficiency) and HA‐CLF (protein interactions). Only in the samples containing both SUP‐GFP and HA‐CLF, HA‐CLF was pull‐down together with anti‐GFP antibody. Molecular mass of the protein ladder is indicated in kilodaltons (KD).
Similar articles
-
Analysis of the Arabidopsis superman allelic series and the interactions with other genes demonstrate developmental robustness and joint specification of male-female boundary, flower meristem termination and carpel compartmentalization.Ann Bot. 2016 Apr;117(5):905-23. doi: 10.1093/aob/mcw023. Epub 2016 Apr 20. Ann Bot. 2016. PMID: 27098089 Free PMC article.
-
Regulation of floral meristem activity through the interaction of AGAMOUS, SUPERMAN, and CLAVATA3 in Arabidopsis.Plant Reprod. 2018 Mar;31(1):89-105. doi: 10.1007/s00497-017-0315-0. Epub 2017 Dec 7. Plant Reprod. 2018. PMID: 29218596
-
Chromatin-mediated feed-forward auxin biosynthesis in floral meristem determinacy.Nat Commun. 2018 Dec 11;9(1):5290. doi: 10.1038/s41467-018-07763-0. Nat Commun. 2018. PMID: 30538233 Free PMC article.
-
Auxin regulation of Arabidopsis flower development involves members of the AINTEGUMENTA-LIKE/PLETHORA (AIL/PLT) family.J Exp Bot. 2011 Jun;62(10):3311-9. doi: 10.1093/jxb/err127. Epub 2011 Apr 21. J Exp Bot. 2011. PMID: 21511900 Review.
-
Epigenetic Mechanisms Are Critical for the Regulation of WUSCHEL Expression in Floral Meristems.Plant Physiol. 2015 Aug;168(4):1189-96. doi: 10.1104/pp.15.00230. Epub 2015 Mar 31. Plant Physiol. 2015. PMID: 25829464 Free PMC article. Review.
Cited by
-
Robust control of floral meristem determinacy by position-specific multifunctions of KNUCKLES.Proc Natl Acad Sci U S A. 2021 Sep 7;118(36):e2102826118. doi: 10.1073/pnas.2102826118. Proc Natl Acad Sci U S A. 2021. PMID: 34462349 Free PMC article.
-
Fine mapping of BnDM1-the gene regulating indeterminate inflorescence in Brassica napus.Theor Appl Genet. 2023 Jun 11;136(7):151. doi: 10.1007/s00122-023-04384-0. Theor Appl Genet. 2023. PMID: 37302112
-
Control of compound leaf patterning by MULTI-PINNATE LEAF1 (MPL1) in chickpea.Nat Commun. 2023 Dec 7;14(1):8088. doi: 10.1038/s41467-023-43975-9. Nat Commun. 2023. PMID: 38062032 Free PMC article.
-
Do Epigenetic Timers Control Petal Development?Front Plant Sci. 2021 Jul 6;12:709360. doi: 10.3389/fpls.2021.709360. eCollection 2021. Front Plant Sci. 2021. PMID: 34295349 Free PMC article. Review.
-
Plant stem cell research is uncovering the secrets of longevity and persistent growth.Plant J. 2021 Apr;106(2):326-335. doi: 10.1111/tpj.15184. Epub 2021 Mar 25. Plant J. 2021. PMID: 33533118 Free PMC article. Review.
References
-
- Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis . Development 112: 1–20 - PubMed
-
- Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM (1992) SUPERMAN, a regulator of floral homeotic genes in Arabidopsis . Development 114: 599–615 - PubMed
-
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 - PubMed
-
- Breuil‐Broyer S, Morel P, de Almeida‐Engler J, Coustham V, Negrutiu I, Trehin C (2004) High‐resolution boundary analysis during Arabidopsis thaliana flower development. Plant J 38: 182–192 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

