Assessment of established techniques to determine developmental and malignant potential of human pluripotent stem cells
- PMID: 29765017
- PMCID: PMC5954055
- DOI: 10.1038/s41467-018-04011-3
Assessment of established techniques to determine developmental and malignant potential of human pluripotent stem cells
Abstract
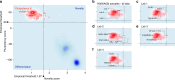
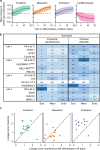
The International Stem Cell Initiative compared several commonly used approaches to assess human pluripotent stem cells (PSC). PluriTest predicts pluripotency through bioinformatic analysis of the transcriptomes of undifferentiated cells, whereas, embryoid body (EB) formation in vitro and teratoma formation in vivo provide direct tests of differentiation. Here we report that EB assays, analyzed after differentiation under neutral conditions and under conditions promoting differentiation to ectoderm, mesoderm, or endoderm lineages, are sufficient to assess the differentiation potential of PSCs. However, teratoma analysis by histologic examination and by TeratoScore, which estimates differential gene expression in each tumor, not only measures differentiation but also allows insight into a PSC's malignant potential. Each of the assays can be used to predict pluripotent differentiation potential but, at this stage of assay development, only the teratoma assay provides an assessment of pluripotency and malignant potential, which are both relevant to the pre-clinical safety assessment of PSCs.
Conflict of interest statement
C.L.M. is a co-founder of Pluriomics bv (from Sept 2017 Ncardia). N.N. is a director and shareholder of hPSC-related companies Stem Cell and Device Laboratory, Inc. and Kyoto Stem Cell Innovation, Inc. He is also a shareholder of ReproCELL, Inc. O.B. is a co-founder and owns equity of LIFE and BRAIN GmbH. S.Y. is a scientific advisor of iPS Academia Japan without salary. K.T. is a member of the scientific advisory board of I Peace Inc. without salary. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

