Detection of Bacteriophage Particles Containing Antibiotic Resistance Genes in the Sputum of Cystic Fibrosis Patients
- PMID: 29765367
- PMCID: PMC5938348
- DOI: 10.3389/fmicb.2018.00856
Detection of Bacteriophage Particles Containing Antibiotic Resistance Genes in the Sputum of Cystic Fibrosis Patients
Abstract
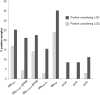
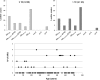
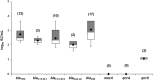
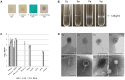
Cystic fibrosis (CF) is a chronic disease in which the bacterial colonization of the lung is linked to an excessive inflammatory response that leads to respiratory failure. The microbiology of CF is complex. Staphylococcus aureus is the first bacterium to colonize the lungs in 30% of pediatric CF patients, and 80% of adult patients develop a chronic Pseudomonas aeruginosa infection, but other microorganisms can also be found. The use of antibiotics is essential to treat the disease, but antibiotic performance is compromised by resistance mechanisms. Among various mechanisms of transfer of antibiotic resistance genes (ARGs), the recently been reported bacteriophages are the least explored in clinical settings. To determine the role of phages in CF as mobile genetic elements (MGEs) carrying ARGs, we evaluated their presence in 71 CF patients. 71 sputum samples taken from these patients were screened for eight ARGs (blaTEM, blaCTX-M-1-group, blaCTX-M-9-group, blaOXA-48, blaVIM, mecA, qnrA, and qnrS) in the bacteriophage DNA fraction. The phages found were also purified and observed by electron microscopy. 32.4% of CF patients harbored ARGs in phage DNA. β-lactamase genes, particularly blaVIM and blaTEM, were the most prevalent and abundant, whereas mecA, qnrA, and qnrS were very rare. Siphoviridae phage particles capable of infecting P. aeruginosa and Klebsiella pneumoniae were detected in CF sputum. Phage particles harboring ARGs were found to be abundant in the lungs of both CF patients and healthy individuals and could contribute to the colonization of multiresistant strains.
Keywords: antibiotic resistance genes; bacteriophages; cystic fibrosis; horizontal gene transfer; sputum.
Figures
References
-
- Anonymous (2000). ISO 10705-2: Water quality. Detection and enumeration of Bacteriophages – part 2: e. Enumeration of Somatic Coliphages. Geneva: Organization for Standardization.
-
- Brown-Jaque M., Calero-Cáceres W., Espinal P., Rodríguez-Navarro J., Miró E., González-López J. J., et al. (2018). Antibiotic resistance genes in phage particles isolated from human feces and induced from clinical bacterial isolates. Int. J. Antimicrob. Agents 51 434–442. 10.1016/j.ijantimicag.2017.11.014 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

