Multi-site evaluation of the LN34 pan-lyssavirus real-time RT-PCR assay for post-mortem rabies diagnostics
- PMID: 29768505
- PMCID: PMC5955534
- DOI: 10.1371/journal.pone.0197074
Multi-site evaluation of the LN34 pan-lyssavirus real-time RT-PCR assay for post-mortem rabies diagnostics
Abstract
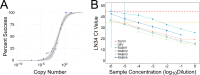
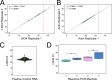
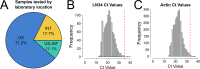
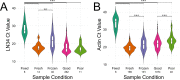
Rabies is a fatal zoonotic disease that requires fast, accurate diagnosis to prevent disease in an exposed individual. The current gold standard for post-mortem diagnosis of human and animal rabies is the direct fluorescent antibody (DFA) test. While the DFA test has proven sensitive and reliable, it requires high quality antibody conjugates, a skilled technician, a fluorescence microscope and diagnostic specimen of sufficient quality. The LN34 pan-lyssavirus real-time RT-PCR assay represents a strong candidate for rabies post-mortem diagnostics due to its ability to detect RNA across the diverse Lyssavirus genus, its high sensitivity, its potential for use with deteriorated tissues, and its simple, easy to implement design. Here, we present data from a multi-site evaluation of the LN34 assay in 14 laboratories. A total of 2,978 samples (1,049 DFA positive) from Africa, the Americas, Asia, Europe, and the Middle East were tested. The LN34 assay exhibited low variability in repeatability and reproducibility studies and was capable of detecting viral RNA in fresh, frozen, archived, deteriorated and formalin-fixed brain tissue. The LN34 assay displayed high diagnostic specificity (99.68%) and sensitivity (99.90%) when compared to the DFA test, and no DFA positive samples were negative by the LN34 assay. The LN34 assay produced definitive findings for 80 samples that were inconclusive or untestable by DFA; 29 were positive. Five samples were inconclusive by the LN34 assay, and only one sample was inconclusive by both tests. Furthermore, use of the LN34 assay led to the identification of one false negative and 11 false positive DFA results. Together, these results demonstrate the reliability and robustness of the LN34 assay and support a role for the LN34 assay in improving rabies diagnostics and surveillance.
Conflict of interest statement
Figures
References
-
- Petersen BW, Rupprecht CE (2011) Human rabies epidemiology and diagnosis In: T S., editor. Non-Flavivirus encephalitis: INTECH Open Access Publisher; doi: 10.5772/21708 - DOI
-
- World Health Organization (2013) WHO Expert Consultation on Rabies: second report. World Health Organization. http://www.who.int/iris/handle/10665/85346 - PubMed
-
- Fooks AR, Cliquet F, Finke S, Freuling C, Hemachudha T, et al. (2017) Rabies. Nat Rev Dis Primers 3: 17091 doi: 10.1038/nrdp.2017.91 - DOI - PubMed
-
- World Organization for Animal Health (OIE) (2017) Rabies (infection with rabies virus) In: Manual of diagnostic tests and vaccines for terrestrial animals. http://www.oie.int/en/international-standard-setting/terrestrial-manual/...
-
- Picard-Meyer E, Barrat J, Cliquet F (2007) Use of filter paper (FTA) technology for sampling, recovery and molecular characterisation of rabies viruses. J Virol Methods 140: 174–182. doi: 10.1016/j.jviromet.2006.11.011 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

