A gene signature that distinguishes conventional and leukemic nonnodal mantle cell lymphoma helps predict outcome
- PMID: 29769262
- PMCID: PMC6071558
- DOI: 10.1182/blood-2018-03-838136
A gene signature that distinguishes conventional and leukemic nonnodal mantle cell lymphoma helps predict outcome
Abstract
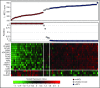
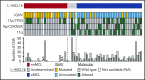
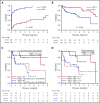
Mantle cell lymphoma (MCL) is an aggressive B-cell malignancy, but some patients have a very indolent evolution. This heterogeneous course is related, in part, to the different biological characteristics of conventional MCL (cMCL) and the distinct subgroup of leukemic nonnodal MCL (nnMCL). Robust criteria to distinguish these MCL subtypes and additional biological parameters that influence their evolution are not well defined. We describe a novel molecular assay that reliably distinguishes cMCL and nnMCL using blood samples. We trained a 16-gene assay (L-MCL16 assay) on the NanoString platform using 19 purified leukemic samples. The locked assay was applied to an independent cohort of 70 MCL patients with leukemic presentation. The assay assigned 37% of cases to nnMCL and 56% to cMCL. nnMCL and cMCL differed in nodal presentation, lactate dehydrogenase, immunoglobulin heavy chain gene mutational status, management options, genomic complexity, and CDKN2A/ATM deletions, but the proportion with 17p/TP53 aberrations was similar in both subgroups. Sequential samples showed that assay prediction was stable over time. nnMCL had a better overall survival (OS) than cMCL (3-year OS 92% vs 69%; P = .006) from the time of diagnosis and longer time to first treatment. Genomic complexity and TP53/CDKN2A aberrations predicted for shorter OS in the entire series and cMCL, whereas only genomic complexity was associated with shorter time to first treatment and OS in nnMCL. In conclusion, the newly developed assay robustly recognizes the 2 molecular subtypes of MCL in leukemic samples. Its combination with genetic alterations improves the prognostic evaluation and may provide useful biological information for management decisions.
© 2018 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: E.C. has received research funding from and has been an expert witness for Gilead Sciences. E.S.J., J.M.C., R.D.G., J.D., G.O., G.W.W., L.M.S., A.R., D.W.S., L.M.S., and E.C. are named inventors on 2 patents filed by the National Cancer Institute: “Methods for selecting and treating lymphoma types” licensed to NanoString Technologies and “Evaluation of mantle cell lymphoma and methods related thereof.” The remaining authors declare no competing financial interests.
Figures



Comment in
-
A tale of two mantle cell lymphomas.Blood. 2018 Jul 26;132(4):347-348. doi: 10.1182/blood-2018-05-853077. Blood. 2018. PMID: 30049731 No abstract available.
References
-
- Campo E, Rule S. Mantle cell lymphoma: evolving management strategies. Blood. 2015;125(1):48-55. - PubMed
-
- Fernàndez V, Salamero O, Espinet B, et al. . Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. 2010;70(4):1408-1418. - PubMed
-
- Orchard J, Garand R, Davis Z, et al. . A subset of t(11;14) lymphoma with mantle cell features displays mutated IgVH genes and includes patients with good prognosis, nonnodal disease. Blood. 2003;101(12):4975-4981. - PubMed
-
- Martin P, Chadburn A, Christos P, et al. . Outcome of deferred initial therapy in mantle-cell lymphoma. J Clin Oncol. 2009;27(8):1209-1213. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

