The chloroplast division protein ARC6 acts to inhibit disassembly of GDP-bound FtsZ2
- PMID: 29769312
- PMCID: PMC6036205
- DOI: 10.1074/jbc.RA117.000999
The chloroplast division protein ARC6 acts to inhibit disassembly of GDP-bound FtsZ2
Abstract
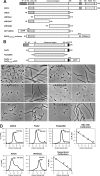
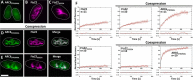
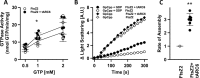
Chloroplasts host photosynthesis and fulfill other metabolic functions that are essential to plant life. They have to divide by binary fission to maintain their numbers throughout cycles of cell division. Chloroplast division is achieved by a complex ring-shaped division machinery located on both the inner (stromal) and the outer (cytosolic) side of the chloroplast envelope. The inner division ring (termed the Z ring) is formed by the assembly of tubulin-like FtsZ1 and FtsZ2 proteins. ARC6 is a key chloroplast division protein that interacts with the Z ring. ARC6 spans the inner envelope membrane, is known to stabilize or maintain the Z ring, and anchors the Z ring to the inner membrane through interaction with FtsZ2. The underlying mechanism of Z ring stabilization is not well-understood. Here, biochemical and structural characterization of ARC6 was conducted using light scattering, sedimentation, and light and transmission EM. The recombinant protein was purified as a dimer. The results indicated that a truncated form of ARC6 (tARC6), representing the stromal portion of ARC6, affects FtsZ2 assembly without forming higher-order structures and exerts its effect via FtsZ2 dynamics. tARC6 prevented GDP-induced FtsZ2 disassembly and caused a significant net increase in FtsZ2 assembly when GDP was present. Single particle analysis and 3D reconstruction were performed to elucidate the structural basis of ARC6 activity. Together, the data reveal that a dimeric form of tARC6 binds to FtsZ2 filaments and does not increase FtsZ polymerization rates but rather inhibits GDP-associated FtsZ2 disassembly.
Keywords: Arabidopsis; GTPase; chloroplast; chloroplast division; cytoskeleton; single particle analysis; structural model.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures




Similar articles
-
In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex.Biochem J. 2008 Jun 1;412(2):367-78. doi: 10.1042/BJ20071354. Biochem J. 2008. PMID: 18284374
-
Roles of Arabidopsis PARC6 in Coordination of the Chloroplast Division Complex and Negative Regulation of FtsZ Assembly.Plant Physiol. 2016 Jan;170(1):250-62. doi: 10.1104/pp.15.01460. Epub 2015 Nov 2. Plant Physiol. 2016. PMID: 26527658 Free PMC article.
-
Chloroplast division protein ARC3 acts on FtsZ2 by preventing filament bundling and enhancing GTPase activity.Biochem J. 2018 Jan 2;475(1):99-115. doi: 10.1042/BCJ20170697. Biochem J. 2018. PMID: 29138260
-
Bacterial Heterologous Expression System for Reconstitution of Chloroplast Inner Division Ring and Evaluation of Its Contributors.Int J Mol Sci. 2018 Feb 11;19(2):544. doi: 10.3390/ijms19020544. Int J Mol Sci. 2018. PMID: 29439474 Free PMC article. Review.
-
Division and dynamic morphology of plastids.Annu Rev Plant Biol. 2014;65:443-72. doi: 10.1146/annurev-arplant-050213-035748. Epub 2014 Jan 22. Annu Rev Plant Biol. 2014. PMID: 24471836 Review.
Cited by
-
Enhanced chloroplast FtsZ-ring constriction by the ARC6-ARC3 module in Arabidopsis.Proc Natl Acad Sci U S A. 2025 Jul;122(26):e2425129122. doi: 10.1073/pnas.2425129122. Epub 2025 Jun 25. Proc Natl Acad Sci U S A. 2025. PMID: 40560617
-
ARC3 Activation by PARC6 Promotes FtsZ-Ring Remodeling at the Chloroplast Division Site.Plant Cell. 2019 Apr;31(4):862-885. doi: 10.1105/tpc.18.00948. Epub 2019 Mar 1. Plant Cell. 2019. PMID: 30824505 Free PMC article.
-
PDV1 and PDV2 Differentially Affect Remodeling and Assembly of the Chloroplast DRP5B Ring.Plant Physiol. 2020 Apr;182(4):1966-1978. doi: 10.1104/pp.19.01490. Epub 2020 Jan 31. Plant Physiol. 2020. PMID: 32005784 Free PMC article.
-
Transcriptomic study of the role of MeFtsZ2-1 in pigment accumulation in cassava leaves.BMC Genomics. 2024 May 7;25(1):448. doi: 10.1186/s12864-024-10165-w. BMC Genomics. 2024. PMID: 38802758 Free PMC article.
References
-
- Martin W., and Kowallik K. V. (1999) Annotated English translation of Mereschkowsky's 1905 paper “Über Natur und Ursprung der Chromatophoren im Pflanzenreiche”. Eur. J. Phycol. 34, 287–296
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

