Differential Responses by Human Respiratory Epithelial Cell Lines to Respiratory Syncytial Virus Reflect Distinct Patterns of Infection Control
- PMID: 29769339
- PMCID: PMC6052282
- DOI: 10.1128/JVI.02202-17
Differential Responses by Human Respiratory Epithelial Cell Lines to Respiratory Syncytial Virus Reflect Distinct Patterns of Infection Control
Abstract
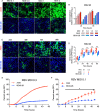
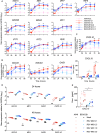
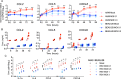
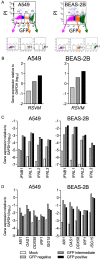
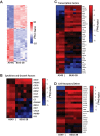
Respiratory syncytial virus (RSV) infects small foci of respiratory epithelial cells via infected droplets. Infection induces expression of type I and III interferons (IFNs) and proinflammatory cytokines, the balance of which may restrict viral replication and affect disease severity. We explored this balance by infecting two respiratory epithelial cell lines with low doses of recombinant RSV expressing green fluorescent protein (rgRSV). A549 cells were highly permissive, whereas BEAS-2B cells restricted infection to individual cells or small foci. After infection, A549 cells expressed higher levels of IFN-β-, IFN-λ-, and NF-κB-inducible proinflammatory cytokines. In contrast, BEAS-2B cells expressed higher levels of antiviral interferon-stimulated genes, pattern recognition receptors, and other signaling intermediaries constitutively and after infection. Transcriptome analysis revealed that constitutive expression of antiviral and proinflammatory genes predicted responses by each cell line. These two cell lines provide a model for elucidating critical mediators of local control of viral infection in respiratory epithelial cells.IMPORTANCE Airway epithelium is both the primary target of and the first defense against respiratory syncytial virus (RSV). Whether RSV replicates and spreads to adjacent epithelial cells depends on the quality of their innate immune responses. A549 and BEAS-2B are alveolar and bronchial epithelial cell lines, respectively, that are often used to study RSV infection. We show that A549 cells are permissive to RSV infection and express genes characteristic of a proinflammatory response. In contrast, BEAS-2B cells restrict infection and express genes characteristic of an antiviral response associated with expression of type I and III interferons. Transcriptome analysis of constitutive gene expression revealed patterns that may predict the response of each cell line to infection. This study suggests that restrictive and permissive cell lines may provide a model for identifying critical mediators of local control of infection and stresses the importance of the constitutive antiviral state for the response to viral challenge.
Keywords: cytokine; cytokines; interferon; interferon regulatory factor (IRF); interferon-stimulated genes; interferons; respiratory epithelial cells; respiratory syncytial virus.
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Figures





References
-
- Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi:10.1016/S0140-6736(10)60206-1. - DOI - PMC - PubMed
-
- Liu P, Jamaluddin M, Li K, Garofalo RP, Casola A, Brasier AR. 2007. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J Virol 81:1401–1411. doi:10.1128/JVI.01740-06. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

