Mxra8 is a receptor for multiple arthritogenic alphaviruses
- PMID: 29769725
- PMCID: PMC5970976
- DOI: 10.1038/s41586-018-0121-3
Mxra8 is a receptor for multiple arthritogenic alphaviruses
Abstract
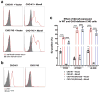
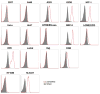
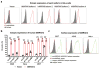
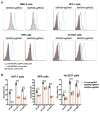
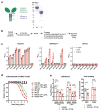
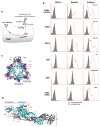
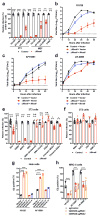
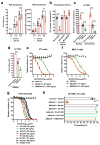
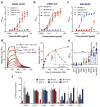
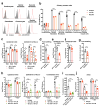
Arthritogenic alphaviruses comprise a group of enveloped RNA viruses that are transmitted to humans by mosquitoes and cause debilitating acute and chronic musculoskeletal disease 1 . The host factors required for alphavirus entry remain poorly characterized 2 . Here we use a genome-wide CRISPR-Cas9-based screen to identify the cell adhesion molecule Mxra8 as an entry mediator for multiple emerging arthritogenic alphaviruses, including chikungunya, Ross River, Mayaro and O'nyong nyong viruses. Gene editing of mouse Mxra8 or human MXRA8 resulted in reduced levels of viral infection of cells and, reciprocally, ectopic expression of these genes resulted in increased infection. Mxra8 bound directly to chikungunya virus particles and enhanced virus attachment and internalization into cells. Consistent with these findings, Mxra8-Fc fusion protein or anti-Mxra8 monoclonal antibodies blocked chikungunya virus infection in multiple cell types, including primary human synovial fibroblasts, osteoblasts, chondrocytes and skeletal muscle cells. Mutagenesis experiments suggest that Mxra8 binds to a surface-exposed region across the A and B domains of chikungunya virus E2 protein, which are a speculated site of attachment. Finally, administration of the Mxra8-Fc protein or anti-Mxra8 blocking antibodies to mice reduced chikungunya and O'nyong nyong virus infection as well as associated foot swelling. Pharmacological targeting of Mxra8 could form a strategy for mitigating infection and disease by multiple arthritogenic alphaviruses.
Figures




Comment in
-
From literature search to vaccine candidate without a lab.Nature. 2018 Jul;559(7715):477. doi: 10.1038/d41586-018-05790-x. Nature. 2018. PMID: 30042540 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

