So many genes, so little time: A practical approach to divergence-time estimation in the genomic era
- PMID: 29772020
- PMCID: PMC5957400
- DOI: 10.1371/journal.pone.0197433
So many genes, so little time: A practical approach to divergence-time estimation in the genomic era
Abstract
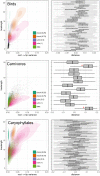
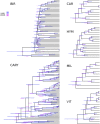
Phylogenomic datasets have been successfully used to address questions involving evolutionary relationships, patterns of genome structure, signatures of selection, and gene and genome duplications. However, despite the recent explosion in genomic and transcriptomic data, the utility of these data sources for efficient divergence-time inference remains unexamined. Phylogenomic datasets pose two distinct problems for divergence-time estimation: (i) the volume of data makes inference of the entire dataset intractable, and (ii) the extent of underlying topological and rate heterogeneity across genes makes model mis-specification a real concern. "Gene shopping", wherein a phylogenomic dataset is winnowed to a set of genes with desirable properties, represents an alternative approach that holds promise in alleviating these issues. We implemented an approach for phylogenomic datasets (available in SortaDate) that filters genes by three criteria: (i) clock-likeness, (ii) reasonable tree length (i.e., discernible information content), and (iii) least topological conflict with a focal species tree (presumed to have already been inferred). Such a winnowing procedure ensures that errors associated with model (both clock and topology) mis-specification are minimized, therefore reducing error in divergence-time estimation. We demonstrated the efficacy of this approach through simulation and applied it to published animal (Aves, Diplopoda, and Hymenoptera) and plant (carnivorous Caryophyllales, broad Caryophyllales, and Vitales) phylogenomic datasets. By quantifying rate heterogeneity across both genes and lineages we found that every empirical dataset examined included genes with clock-like, or nearly clock-like, behavior. Moreover, many datasets had genes that were clock-like, exhibited reasonable evolutionary rates, and were mostly compatible with the species tree. We identified overlap in age estimates when analyzing these filtered genes under strict clock and uncorrelated lognormal (UCLN) models. However, this overlap was often due to imprecise estimates from the UCLN model. We find that "gene shopping" can be an efficient approach to divergence-time inference for phylogenomic datasets that may otherwise be characterized by extensive gene tree heterogeneity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Smith SA, Beaulieu JM, Donoghue MJ. An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proceedings of the National Academy of Sciences. 2010;107(13):5897–5902. doi: 10.1073/pnas.1001225107 - DOI - PMC - PubMed
-
- Dornburg A, Brandley MC, McGowen MR, Near TJ. Relaxed Clocks and Inferences of Heterogeneous Patterns of Nucleotide Substitution and Divergence Time Estimates across Whales and Dolphins (Mammalia: Cetacea). Molecular Biology and Evolution. 2012;29(2):721–736. doi: 10.1093/molbev/msr228 - DOI - PubMed
-
- Parham JF, Donoghue PCJ, Bell CJ, Calway TD, Head JJ, Holroyd PA, et al. Best Practices for Justifying Fossil Calibrations. Systematic Biology. 2012;61(2):346–359. doi: 10.1093/sysbio/syr107 - DOI - PMC - PubMed
-
- Heath TA, Moore BR. Bayesian inference of species divergence times In: Chen MH, Kuo L, Lewis PO, editors. Bayesian Phylogenetics: Methods Algorithms, and Applications. Boca Raton, Florida: CRC Press; 2014. p. 277–318.
-
- Beaulieu JM, O’Meara BC, Crane P, Donoghue MJ. Heterogeneous Rates of Molecular Evolution and Diversification Could Explain the Triassic Age Estimate for Angiosperms. Systematic Biology. 2015;64(5):869–878. doi: 10.1093/sysbio/syv027 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

