Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin
- PMID: 29773078
- PMCID: PMC5956843
- DOI: 10.1186/s40168-018-0470-z
Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin
Abstract
Background: Taxonomic classification of marker-gene sequences is an important step in microbiome analysis.
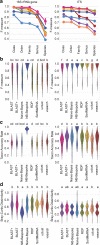
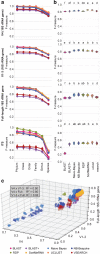
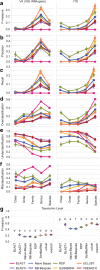
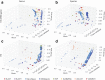
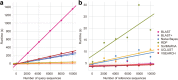
Results: We present q2-feature-classifier ( https://github.com/qiime2/q2-feature-classifier ), a QIIME 2 plugin containing several novel machine-learning and alignment-based methods for taxonomy classification. We evaluated and optimized several commonly used classification methods implemented in QIIME 1 (RDP, BLAST, UCLUST, and SortMeRNA) and several new methods implemented in QIIME 2 (a scikit-learn naive Bayes machine-learning classifier, and alignment-based taxonomy consensus methods based on VSEARCH, and BLAST+) for classification of bacterial 16S rRNA and fungal ITS marker-gene amplicon sequence data. The naive-Bayes, BLAST+-based, and VSEARCH-based classifiers implemented in QIIME 2 meet or exceed the species-level accuracy of other commonly used methods designed for classification of marker gene sequences that were evaluated in this work. These evaluations, based on 19 mock communities and error-free sequence simulations, including classification of simulated "novel" marker-gene sequences, are available in our extensible benchmarking framework, tax-credit ( https://github.com/caporaso-lab/tax-credit-data ).
Conclusions: Our results illustrate the importance of parameter tuning for optimizing classifier performance, and we make recommendations regarding parameter choices for these classifiers under a range of standard operating conditions. q2-feature-classifier and tax-credit are both free, open-source, BSD-licensed packages available on GitHub.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

