PTAP motif duplication in the p6 Gag protein confers a replication advantage on HIV-1 subtype C
- PMID: 29773649
- PMCID: PMC6066301
- DOI: 10.1074/jbc.M117.815829
PTAP motif duplication in the p6 Gag protein confers a replication advantage on HIV-1 subtype C
Abstract
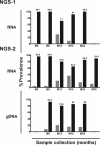
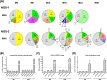
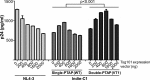
HIV-1 subtype C (HIV-1C) may duplicate longer amino acid stretches in the p6 Gag protein, leading to the creation of an additional Pro-Thr/Ser-Ala-Pro (PTAP) motif necessary for viral packaging. However, the biological significance of a duplication of the PTAP motif for HIV-1 replication and pathogenesis has not been experimentally validated. In a longitudinal study of two different clinical cohorts of select HIV-1 seropositive, drug-naive individuals from India, we found that 8 of 50 of these individuals harbored a mixed infection of viral strains discordant for the PTAP duplication. Conventional and next-generation sequencing of six primary viral quasispecies at multiple time points disclosed that in a mixed infection, the viral strains containing the PTAP duplication dominated the infection. The dominance of the double-PTAP viral strains over a genetically similar single-PTAP viral clone was confirmed in viral proliferation and pairwise competition assays. Of note, in the proximity ligation assay, double-PTAP Gag proteins exhibited a significantly enhanced interaction with the host protein tumor susceptibility gene 101 (Tsg101). Moreover, Tsg101 overexpression resulted in a biphasic effect on HIV-1C proliferation, an enhanced effect at low concentration and an inhibitory effect only at higher concentrations, unlike a uniformly inhibitory effect on subtype B strains. In summary, our results indicate that the duplication of the PTAP motif in the p6 Gag protein enhances the replication fitness of HIV-1C by engaging the Tsg101 host protein with a higher affinity. Our results have implications for HIV-1 pathogenesis, especially of HIV-1C.
Keywords: Gag; HIV-1C; NGS; PTAP duplication; Replication advantage; human immunodeficiency virus (HIV); infectious disease; viral DNA; viral protein; viral replication.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- Bachu M., Yalla S., Asokan M., Verma A., Neogi U., Sharma S., Murali R. V., Mukthey A. B., Bhatt R., Chatterjee S., Rajan R. E., Cheedarla N., Yadavalli V. S., Mahadevan A., Shankar S. K., et al. (2012) Multiple NF-κB sites in HIV-1 subtype C long terminal repeat confer superior magnitude of transcription and thereby the enhanced viral predominance. J. Biol. Chem. 287, 44714–44735 10.1074/jbc.M112.397158 - DOI - PMC - PubMed
-
- Ramírez d A., Soriano V., Alcamil J., and Holguín A. (2006) New findings on transcription regulation across different HIV-1 subtypes. AIDS Rev. 8, 9–16 - PubMed
-
- De Baar M. P., De Ronde A., Berkhout B., Cornelissen M., Van Der Horn K. H., Van Der Schoot A. M., De Wolf F., Lukashov V. V., and Goudsmit J. (2000) Subtype-specific sequence variation of the HIV type 1 long terminal repeat and primer-binding site. AIDS Res. Hum. Retroviruses 16, 499–504 10.1089/088922200309160 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

