Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes
- PMID: 29773668
- PMCID: PMC6442475
- DOI: 10.1126/science.aar4142
Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes
Abstract
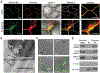
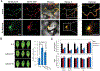
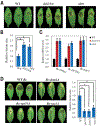
Some pathogens and pests deliver small RNAs (sRNAs) into host cells to suppress host immunity. Conversely, hosts also transfer sRNAs into pathogens and pests to inhibit their virulence. Although sRNA trafficking has been observed in a wide variety of interactions, how sRNAs are transferred, especially from hosts to pathogens and pests, is still unknown. Here, we show that host Arabidopsis cells secrete exosome-like extracellular vesicles to deliver sRNAs into fungal pathogen Botrytis cinerea These sRNA-containing vesicles accumulate at the infection sites and are taken up by the fungal cells. Transferred host sRNAs induce silencing of fungal genes critical for pathogenicity. Thus, Arabidopsis has adapted exosome-mediated cross-kingdom RNA interference as part of its immune responses during the evolutionary arms race with the pathogen.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures

Comment in
-
Targeting microbial pathogens.Science. 2018 Jun 8;360(6393):1070-1071. doi: 10.1126/science.aat9343. Science. 2018. PMID: 29880672 No abstract available.
-
The Trojan Horse of the Plant Kingdom.Cell Host Microbe. 2018 Jul 11;24(1):1-3. doi: 10.1016/j.chom.2018.06.015. Cell Host Microbe. 2018. PMID: 30001514
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

