The relative resistance of children to sepsis mortality: from pathways to drug candidates
- PMID: 29773677
- PMCID: PMC5974511
- DOI: 10.15252/msb.20177998
The relative resistance of children to sepsis mortality: from pathways to drug candidates
Abstract
Attempts to develop drugs that address sepsis based on leads developed in animal models have failed. We sought to identify leads based on human data by exploiting a natural experiment: the relative resistance of children to mortality from severe infections and sepsis. Using public datasets, we identified key differences in pathway activity (Pathprint) in blood transcriptome profiles of septic adults and children. To find drugs that could promote beneficial (child) pathways or inhibit harmful (adult) ones, we built an in silico pathway drug network (PDN) using expression correlation between drug, disease, and pathway gene signatures across 58,475 microarrays. Specific pathway clusters from children or adults were assessed for correlation with drug-based signatures. Validation by literature curation and by direct testing in an endotoxemia model of murine sepsis of the most correlated drug candidates demonstrated that the Pathprint-PDN methodology is more effective at generating positive drug leads than gene-level methods (e.g., CMap). Pathway-centric Pathprint-PDN is a powerful new way to identify drug candidates for intervention against sepsis and provides direct insight into pathways that may determine survival.
Keywords: connectivity map; drug discovery; pathways; sepsis.
© 2018 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
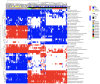

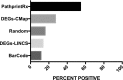
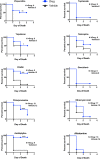

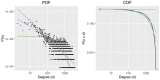
Comment in
-
Learning lessons in sepsis from the children.Mol Syst Biol. 2018 May 17;14(5):e8335. doi: 10.15252/msb.20188335. Mol Syst Biol. 2018. PMID: 29773678 Free PMC article.
References
-
- Ahn S‐H, Tsalik EL, Cyr DD, Zhang Y, van Velkinburgh JC, Langley RJ, Glickman SW, Cairns CB, Zaas AK, Rivers EP, Otero RM, Veldman T, Kingsmore SF, Lucas J, Woods CW, Ginsburg GS, Fowler J, Vance G (2013) Gene expression‐based classifiers identify Staphylococcus aureus infection in mice and humans. PLoS One 8: e48979 - PMC - PubMed
-
- Albert R (2005) Scale‐free networks in cell biology. J Cell Sci 118: 4947–4957 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

