NEDD9 promotes oncogenic signaling, a stem/mesenchymal gene signature, and aggressive ovarian cancer growth in mice
- PMID: 29773902
- PMCID: PMC6119087
- DOI: 10.1038/s41388-018-0296-y
NEDD9 promotes oncogenic signaling, a stem/mesenchymal gene signature, and aggressive ovarian cancer growth in mice
Abstract
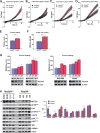
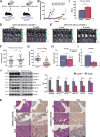
Neural precursor cell expressed, developmentally downregulated 9 (NEDD9) supports oncogenic signaling in a number of solid and hematologic tumors. Little is known about the role of NEDD9 in ovarian carcinoma (OC), but available data suggest elevated mRNA and protein expression in advanced stage high-grade cancers. We used a transgenic MISIIR-TAg mouse OC model combined with genetic ablation of Nedd9 to investigate its action in the development and progression of OC. A Nedd9-/- genotype delayed tumor growth rate, reduced incidence of ascites, and reduced expression and activation of signaling proteins including SRC, STAT3, E-cadherin, and AURKA. Cell lines established from MISIIR-TAg;Nedd9-/- and MISIIR-TAg;Nedd9+/+ mice exhibited altered migration and invasion. Growth of these cells in a syngeneic allograft model indicated that systemic Nedd9 loss in the microenvironment had little impact on tumor allograft growth, but in a Nedd9 wild-type background Nedd9-/- allografts exhibited significantly reduced growth, dissemination, and oncogenic signaling compared to Nedd9+/+ allografts. Gene expression analysis revealed that Nedd9+/+ tumors exhibited more mesenchymal "stem-like" transcriptional program, including increased expression of Aldh1a1 and Aldh1a2. Conversely, loss of Nedd9 resulted in increased expression of differentiation genes, including fallopian tube markers Foxj1, Ovgp1, and Pax8. Collectively, these data suggest that tumor cell-intrinsic Nedd9 expression promotes OC development and progression by broad induction of oncogenic protein signaling and stem/mesenchymal gene expression.
Conflict of interest statement
Figures




References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A Cancer Journal for Clinicians. 2016;66(1):7–30. - PubMed
-
- McPherson A, Roth A, Laks E, Masud T, Bashashati A, Zhang AW, et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat Genet. 2016;48(7):758–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

