The Invasive Brazilian Pepper Tree (Schinus terebinthifolius) Is Colonized by a Root Microbiome Enriched With Alphaproteobacteria and Unclassified Spartobacteria
- PMID: 29774018
- PMCID: PMC5943492
- DOI: 10.3389/fmicb.2018.00876
The Invasive Brazilian Pepper Tree (Schinus terebinthifolius) Is Colonized by a Root Microbiome Enriched With Alphaproteobacteria and Unclassified Spartobacteria
Abstract
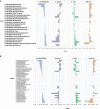
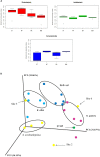
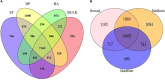
Little is known about the rhizosphere microbiome of the Brazilian pepper tree (BP) - a noxious category 1 invasive plant inducing an enormous economic and ecological toll in Florida. Some invasive plants have been shown to drastically change the soil microbiome compared to other native plants. The rhizobacteria community structure of BP, two Florida native plants (Hamelia patens and Bidens alba) and bulk soils were characterized across six geographical sites. Although all 19 well-known and 10 poorly described phyla were observed in all plant rhizospheres, BP contained the least total bacterial abundance (OTUs) with a distinct bacteria community structure and clustering patterns differing significantly (pCOA and PERMANOVA) from the natives and bulk soil. The BP rhizosphere community contained the highest overall Proteobacteria diversity (Shannon's diversity 3.25) in spite of a twofold reduction in richness of the Gammaproteobacteria. Remarkably, the invasive BP rhizosphere was highly enriched with Alphaproteobacteria, dominated by Rhizobiales, including Rhodoplanes and Bradyrhizobiaceae. Also, the relative abundance of Spartobacteria under BP rhizosphere was more than twice that of native plants and bulk soil; featuring unique members of the family Chthoniobacteraceae (DA101 genus). The trend was different for the family Pedosphaerae in the phylum Verrucomicrobia where the abundance declined under BP (26%) compared to (33-66%) for the H. patens native plant and bulk soil. BP shared the lowest number of unique phylotypes with bulk soil (146) compared to the other native plants with bulk soil (B. alba - 222, H. patens - 520) suggestive of its capacity to overcome biotic resistance. Although there were no specific biomarkers found, taken together, our data suggests that the occurrence of key bacteria groups across multiple taxonomic ranks provides a somewhat consistent profile of the invasive BP rhizo-community. Furthermore, based on the observed prevalence of a bacteria group (Spartobacteria - Chthoniobacteraceae - DA101); we propose that they have a possible role in BP biology. Our results emphasize the need to further investigate the potential value of "unique phylotypes" in the rhizosphere relative to bulk soil as an ecological tool for monitoring plant-cover/invasion history; or even detecting exotic plants with invasion tendencies.
Keywords: Brazilian pepper tree; Spartobacteria; biotic resistance; invasive plant; microbiome; rhizosphere.
Figures

Similar articles
-
Arbuscular and Ectomycorrhizal Fungi Associated with the Invasive Brazilian Pepper Tree (Schinus terebinthifolius) and Two Native Plants in South Florida.Front Microbiol. 2017 Apr 20;8:665. doi: 10.3389/fmicb.2017.00665. eCollection 2017. Front Microbiol. 2017. PMID: 28473811 Free PMC article.
-
Emerging Insights on Brazilian Pepper Tree (Schinus terebinthifolius) Invasion: The Potential Role of Soil Microorganisms.Front Plant Sci. 2016 May 24;7:712. doi: 10.3389/fpls.2016.00712. eCollection 2016. Front Plant Sci. 2016. PMID: 27252726 Free PMC article. Review.
-
Restoration with pioneer plants changes soil properties and remodels the diversity and structure of bacterial communities in rhizosphere and bulk soil of copper mine tailings in Jiangxi Province, China.Environ Sci Pollut Res Int. 2018 Aug;25(22):22106-22119. doi: 10.1007/s11356-018-2244-3. Epub 2018 May 25. Environ Sci Pollut Res Int. 2018. PMID: 29802615
-
Analysis of the community composition and bacterial diversity of the rhizosphere microbiome across different plant taxa.Microbiologyopen. 2019 Jun;8(6):e00762. doi: 10.1002/mbo3.762. Epub 2018 Nov 22. Microbiologyopen. 2019. PMID: 30565881 Free PMC article.
-
Soil bacterial communities of paddy are dependent on root compartment niches but independent of growth stages from Mollisols of Northeast China.Front Microbiol. 2023 Apr 14;14:1170611. doi: 10.3389/fmicb.2023.1170611. eCollection 2023. Front Microbiol. 2023. PMID: 37125155 Free PMC article.
Cited by
-
Significant changes in soil microbial community structure and metabolic function after Mikania micrantha invasion.Sci Rep. 2023 Jan 20;13(1):1141. doi: 10.1038/s41598-023-27851-6. Sci Rep. 2023. PMID: 36670134 Free PMC article.
-
Dynamics of Soil Bacterial and Fungal Communities During the Secondary Succession Following Swidden Agriculture IN Lowland Forests.Front Microbiol. 2021 Jun 7;12:676251. doi: 10.3389/fmicb.2021.676251. eCollection 2021. Front Microbiol. 2021. PMID: 34163452 Free PMC article.
-
The Rhizosphere Microbiome of Mikania micrantha Provides Insight Into Adaptation and Invasion.Front Microbiol. 2020 Jul 7;11:1462. doi: 10.3389/fmicb.2020.01462. eCollection 2020. Front Microbiol. 2020. PMID: 32733410 Free PMC article.
-
Exploring the diversity and potential functional characteristics of microbiota associated with different compartments of Schisandra chinensis.Front Microbiol. 2024 Jun 13;15:1419943. doi: 10.3389/fmicb.2024.1419943. eCollection 2024. Front Microbiol. 2024. PMID: 38939187 Free PMC article.
-
Cassava/peanut intercropping improves soil quality via rhizospheric microbes increased available nitrogen contents.BMC Biotechnol. 2020 Feb 28;20(1):13. doi: 10.1186/s12896-020-00606-1. BMC Biotechnol. 2020. PMID: 32111197 Free PMC article.
References
-
- Badri D. V., Chaparro J. M., Shen Q., Vivanco J. M. (2013). Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288 4502–4512. 10.1074/jbc.M112.433300 - DOI - PMC - PubMed
-
- Batten K. M., Scow K. M., Davies K. F., Harrison S. P. (2006). Two invasive plants alter soil microbial community composition in serpentine grasslands. Biol. Invasions 8 217–230. 10.1007/s10530-004-3856-8 - DOI
-
- Bennett J. A., Stotz G. C., Cahill J. F., Bruun H. H. (2014). Patterns of phylogenetic diversity are linked to invasion impacts, not invasion resistance, in a native grassland. J. Veg. Sci. 25 1315–1326. 10.1111/jvs.12199 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

