Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma
- PMID: 29774077
- PMCID: PMC5957011
- DOI: 10.7150/thno.23012
Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma
Abstract
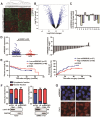
Long noncoding RNAs (lncRNAs) have been associated with hepatocellular carcinoma (HCC), but the underlying molecular mechanisms of their specific association with hepatocarcinogenesis have not been fully explored. Methods: miR503HG was identified by microarray and validated by real-time PCR. Survival analysis was evaluated using the Kaplan-Meier method and assessed using the log-rank test. In vitro and in vivo assays were preformed to explore the biological effects of miR503HG in HCC cells. The interaction of miR503HG with HNRNPA2B1 was identified by RNA pull-down and RNA immunoprecipitation. Expression of HNRNPA2B1 was examined by western blotting, immunofluorescence and immunohistochemical analyses, while HNRNPA2B1 ubiquitination was detected by immunoprecipitation. Results: We have identified 713 differentially expressed lncRNAs in 12 pairs of HCC tissues compared with corresponding noncancerous liver tissues. One of these lncRNAs, miR503HG, the host gene of miR503, is markedly decreased in HCC. Expression level of miR503HG is significantly associated with the time to recurrence and overall survival and is an independent risk factor for recurrence and survival. Enhanced expression of miR503HG could noticeably inhibit HCC invasion and metastasis in vitro and in vivo. Further investigation suggested that miR503HG could specifically interact with the heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1). miR503HG promoted HNRNPA2B1 degradation via the ubiquitin-proteasome pathway, which reduced the stability of p52 and p65 mRNA, and simultaneously suppressed the NF-κB signaling pathway in HCC cells. In addition, miR503HG can function synergistically with miR503 to inhibit HCC migration. Conclusion: Our findings support a role for miR503HG in tumor recurrence risk and survival prediction in HCC patients. We demonstrate a novel mechanism by which miR503HG inhibits the NF-κB signaling pathway and exerts its metastatic tumor suppression function through modulating the ubiquitination status of HNRNPA2B1.
Keywords: hepatocellular carcinoma; lncRNA; metastasis; miR503HG; prognosis.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
-
- Aldrighetti L, Pulitano C, Catena M, Arru M, Guzzetti E, Halliday J, Ferla G. Liver resection with portal vein thrombectomy for hepatocellular carcinoma with vascular invasion. Ann Surg Oncol. 2009;16:1254. - PubMed
-
- Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M, Ge C. et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12:390–99. - PubMed
-
- Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, Yan M. et al. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846–56. - PubMed
-
- Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen T, Chen Z. et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. 2015;62:1132–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

